The 2013 article is careful to define what is meant by random in the modern synthesis:
“I will use the definition that the changes are assumed to be random with respect to physiological function and could not therefore be influenced by such function or by functional changes in response to the environment. This is the assumption that excludes the phenotype from in any way influencing or guiding genetic change.”
Some have criticised the article and the videos on the grounds that the modern synthesis does not require DNA change to be equally likely everywhere in the genome. The article does not make that claim. On the contrary it states:
“The concept of a purely random event is not easy to define. The physico-chemical nature of biological molecules will, in any case, ensure that some changes are more likely to happen than others.”
Of course the modern synthesis can accept non-random change. That has been known ever since the identification of genome hotspots. What is completely contrary to the spirit of the modern synthesis is targeted DNA change that is functionally significant. We now have examples of that kind of change. To read more on those examples see the answer to the question on the relevance to physiology.
The relevant part of the IUPS2013 lecture starts at 7 minutes with the phrase “It is important to ask the question what we mean by random” followed at 7:18 by “rather by whether the changes are functionally relevant” before quoting the paragraph from the article shown above. The article and the lecture could not be clearer. See also Randomness and Function.
It is remarkable how often the same unthinking criticism of the article and the lecture turn up on blog websites. What this shows is that the writers have not taken the time to read the article or listen carefully to the video lectures. Having missed the target on this matter, the same bloggers usually go on to the further false accusation that I claim to have disproved “the theory of evolution”. Anyone who reads the article or watches the videos would find that laughable, so why do they do it? For some strange reason defenders of neo-Darwinism on blogs seem to think that anyone who questions neo-Darwinism is questioning the existence of evolution. The article clearly states:
“In some respects, my article returns to a more nuanced, less dogmatic view of evolutionary theory (see also Müller, 2007; Mesoudi et al. 2013), which is much more in keeping with the spirit of Darwin’s own ideas than is the Neo-Darwinist view.”
As I write in another answer (Answers-dogmatism.html): “It is perfectly possible to defend the virtual certainty that life has evolved while debating in the usual argumentative scientific way the uncertainties surrounding the question of mechanisms.” That is where the real debate is taking place: what are the relative contributions of the various mechanisms to the evolutionary process and the speed with which it has occurred (see Speed of Evolution).
Central Dogma
The central dogma of molecular biology seemed to confirm the Weismann barrier idea. But it doesn’t. The dogma was misinterpreted to mean that information could not pass from the organism and environment to the genome. To quote The Selfish Gene, genes are “sealed off from the outside world.” This is simply incorrect.
“The mechanisms of transposable elements illustrate one of the important breaks with the central dogma of molecular biology. Retrotransposons are DNA sequences that are first copied as RNA sequences, which are then inserted back into a different part of the genome using reverse transcriptase. DNA transposons may use a cut-and-paste mechanism that does not require an RNA intermediate. As Beurton et al.(2008) comment, ‘it seems that a cell’s enzymes are capable of actively manipulating DNA to do this or that. A genome consists largely of semi-stable genetic elements that may be rearranged or even moved around in the genome thus modifying the information content of DNA.’ The central dogma of the 1950s, as a general principle of biology, has therefore been progressively undermined until it has become useless as support for the Modern Synthesis (Werner, 2005; Mattick, 2007; Shapiro, 2009) or indeed as an accurate description of what happens in cells. As Mattick (2012) says, ‘the belief that the soma and germ line do not communicate is patently incorrect.’”
Beurton PJ, Falk R & Rheinberger H.-J. (2008). The Concept of the Gene in Development and Evolution: Historical and Epistemological Perspectives. Cambridge University Press, Cambridge, UK.
Mattick JS (2007). Deconstructing the dogma: a new view of the evolution and genetic programming of complex organisms. Ann N Y Acad Sci 1178, 29–46. http://www.ncbi.nlm.nih.gov/pubmed/19845626
Mattick JS (2012). Rocking the foundations of molecular genetics. Proc Natl Acad Sci USA109, 16400–16401 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3478605/
Shapiro JA (2009). Revisiting the central dogma in the 21st century. Ann N Y Acad Sci 1178, 6–28. http://www.ncbi.nlm.nih.gov/pubmed/19845625
Werner E (2005). Genome semantics, in silico multicellular systems and the Central Dogma. FEBS Lett 579, 1779–1782. http://www.ncbi.nlm.nih.gov/pubmed/15763551
It is important to note that Crick’s original statement of the dogma (made in 1958, and repeated in 1970) was qualified in a very important respect:
“The central dogma of molecular biology deals with the detailed residue-by-residue transfer of sequential information. It states that such information cannot be transferred back from protein to either protein or nucleic acid.”
I have italicised “such information” to highlight this important qualification. The statement does not exclude control information passing from cells to the genome. This must happen to enable the same genome to be used to generate many different cell types.
The Origin of Species?
Despite the title of Darwin’s famous book, we cannot be sure to what extent natural selection acting on gradual accumulation of small mutations has accounted for the development of new species. Many of the examples that are often quoted as demonstrating that this is the main source of new species show that new species have arisen in new niches, e.g. by geographic isolation as in the case of the Galapagos island tortoises and finches. But this evidence, by itself, does not establish what the mechanism may have been. Other interpretations would also be possible.
Darwin’s idea of natural selection was in part based on observing the effects of artificial selection, producing many different varieties of dogs, cats, fish etc. But it is important to note that thousands of years of this kind of selection has produced new varieties, not fully-fledged new species as defined by inability to interbreed. At best, this demonstrates selection’s ability to produce incipient development of new species.
Perhaps the best example of species development is the variety of greenish warblers, forming what is called a ring species around the Himalayas. See http://www.zoology.ubc.ca/~irwin/greenishwarblers.html
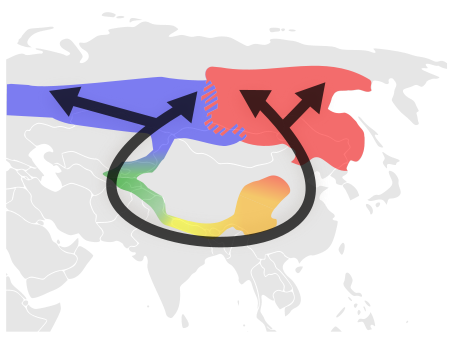
Presumed evolution of Greenish Warblers around the Himalayas. The colours indicate the subspecies: Yellow: P. t. trochiloides; Orange: P. t. obscuratus; Red: P. t. plumbeitarsus; Green: P. t. “ludlowi”; Blue: P. t. viridanus.
From Wikimedia Commons. Wikipedia entry on Greenish Warblers
North of the Himalayas there are two varieties (subspecies?) that co-exist but do not interbreed. But they are each connected to varieties with which they do interbreed stretching around the Himalayas to join the ring together in the south. Ticehurst proposed in 1938 that the greenish warblers started in the south, then slowly evolved as they spread around the Himalayas to the west and east, to eventually meet in the north when they had become different species. Genetic data are consistent with his hypothesis. This is perhaps the best example we have of this process, which itself is rare. Note however that this evidence supports Ticehurst’s hypothesis about the historical development, since the two northern forms are the most distinct genetically, but it does not in itself prove the precise mechanism. It does not, for example, exclude a Waddington-type influence of the environment on the selection of combinations of genes, or any other process by which a Lamarckian process becomes assimilated into the genome. Nor does it exclude natural genetic engineering. This is a difficulty with any study of existing genetic variations to infer the process by which those variations originally arose. The role of natural genetic engineering could be determined when we have genome sequences from most or all of the varieties.
By contrast, hybridisation in plants readily generates new species, and the process of symbiogenesis must also have done so. Natural genetic engineering could also do so if it involves DNA shuffling that removes reproductive compatibility.
The important message here is not that the generation of new species by natural selection working on random mutations is impossible, but rather that the examples often quoted to prove that it happened are not so clear cut when they are examined closely. We have to distinguish evidence about the historical development of new species from evidence about the precise mechanisms by which this happened.
What happened to junk DNA?
Isn’t a lot of DNA “selfish”, “parasitic”?
One of the standard defences of the selfish gene view is based on the discovery that, in humans and many other organisms, only a few per cent of the genome codes for proteins and can therefore be classified as ‘genes’ in the usual sense. The rest was described as ‘junk’ DNA, the ultimate example of ‘selfishness’ since it was seen as DNA ‘hitching a ride’ with no function, a bit like a virus that has become permanently resident in the body. The strong implication is that this discovery favours the selfish gene view.
I think that is a confusing way of viewing genomes. There are several ways in which the confusion can be unravelled.
The words ‘selfish’, ‘junk’, etc. are, of course, metaphors. More importantly they are empirically empty metaphors when applied to sequences of DNA. No conceivable experiment could validate or invalidate them. The reasons are fully explained in Noble (2011). See also the answer to ‘what is wrong with The Selfish Gene?’ in this document.
That the metaphors are empty, however, does not mean that they have no impact. On the contrary, they have had, and still have, very persuasive impact on the way in which many people think about biology. The Selfish Gene sells in millions of copies. Its impact extends way beyond biology, into economics, politics and business studies.
A more persuasive counter-argument is therefore needed, and what might persuade most people is experimental evidence favouring a different view. Fortunately, recent experimental work has provided us with precisely that. The more we examine non-protein-coding DNA the more evidence we find that an overwhelming 80% is transcribed to form RNAs, and that around 20% are already known to have function (http://www.genome.gov/10005107).
Mobile genetic elements have also been characterised as “ selfish” and “parasitic”. Yet they account for nearly 20% of “all conserved (i.e. positively selected) differences between eutherian mammals and marsupials (Lindblad et al , 2011).” For further debate on these issues the reader is referred to the series of articles (July 2013) in Physics of Life Reviews.
What are the implications for the speed of evolutionary change?
What are the implications for the speed of evolutionary change?
Some of the problems in evolutionary theory concern the speed of speciation. It is clear from the geological record that this has not always been smooth. One of the reasons why Cuvier disagreed with Lamarck in the early nineteenth century is that he was able to discredit Lamarck’s idea of the transformation of species by pointing to the fact that the fossil evidence, as it was known then, was also consistent with multiple periods of creation. The record was that patchy, and it was also clear that some species remained essentially unchanged for very long periods of time (stasis). In the twentieth century, with much more evidence to consider, Eldredge and Gould in 1971 proposed the theory of punctuated equilibrium to account for the fact that most fossil species show long periods of stasis, and that rapid (on a geological time scale) changes occurred more rarely and were important periods of speciation. The difference between their theory and that of Cuvier was that Eldredge and Gould were simply proposing that evolutionary change from a common ancestor does not happen at a constant speed, whereas Cuvier interpreted the evidence to show that there had been multiple creations.
Darwin also realised that evolutionary change could not always have been smooth. In the fourth edition of The Origin of Species he wrote “the periods during which species have undergone modification, though long as measured in years, have probably been short in comparison with the periods during which they retain the same form.” This is, in essence, Eldredge and Gould’s idea of punctuated equilibrium.
There has been much argument about what precisely is meant by ‘punctuated’. A gradual change over a hundred thousand or a million or even a few million years will appear rapid on a geological time scale of hundreds of millions of years. The Cambrian explosion that occurred over 500 million years ago is a good example. Within just 20 million years all the phyla in existence today had developed. The standard response to these kinds of theories has therefore been that they are entirely consistent with neo-darwinism. Changes in selection pressure due to environmental changes or geographic distribution, and the occasional catastrophic environmental change, might account for the observed variations in speed of change without supposing additional mechanisms of change.
The new evidence from work on symbiogenesis, the various forms of inheritance of acquired characteristics, genetic assimilation, natural genetic engineering, including genome change and reorganisation over and above the accumulation of chance mutations, changes the situation in more fundamental ways that require either extensions of or replacement of the modern synthesis. These mechanisms resemble punctuated equilibrium theories in proposing that evolution can occur in jumps. Since these can be very sudden indeed it is best to use a word different from ‘punctuated’ to avoid confusion with Eldredge and Gould’s theory. ‘Saltatory’ means jumping. The new mechanisms produce saltations of various kinds:
Symbiogenesis is the fusion of two species. The best established example of this is the bacterial origin of mitochondria and chloroplasts and, perhaps, other organelles. Clearly, this process is the ultimate in ‘saltation’. It depends on processes of cellular ingestion that are natural in the feeding activity of unicellular organisms and, on an evolutionary timescale, it is therefore very rapid indeed. Of course, subsequent changes can then also occur more slowly. We know, for example, that some of the ingested DNA in what became organelles eventually moved to the nucleus in eukaryotes.
See the film by Lynn Margulis on
http://www.voicesfromoxford.com/video/Homage-to-Darwin-part-2/63

Endosymbiosis: Homage to Lynn Margulis, a painting by Shoshanah Dubineer, occupies a hallway in the Morrill Science Center at the University of Massachusetts, Amherst, where Margulis was a professor until her death in 2011. Margulis maintained that genetic variation emerges primarily through symbiosis, not through competition.Click edit button to change this text.
Image courtesy of the artist, http://www.cybermuse.com
See also Evolution’s Other Narrative
Natural genetic engineering could also occur within a single generation. Reorganisations of genomes involving duplications, deletions and insertions of long sequences, would be essentially instantaneous on a geological timescale. Defenders of the modern synthesis have argued that speciation due to such changes, and symbiogenesis, should not be classified as punctuated equilibrium. That is correct in the sense that it was not what Eldridge and Gould had in mind. But so far as timescale is concerned such changes would be saltatory in the ordinary sense of the word. They would be even more sudden than the punctuations proposed by Eldredge and Gould.
Genetic assimilation can also occur rapidly. Waddington’s mid-twentieth century experiments showed that an induced acquired characteristic in fruit flies could become permanent (assimilated) within fourteen generations. This must have represented the time required for selection for an induced characteristic to bring together in a single genome all the relevant alleles for that characteristic to be passed on to subsequent generations without the inducing environmental stimulus. Waddington coined the term ‘epigenetics’ to describe his discovery. Today, epigenetics usually refers to genome and chromatin marking.
Inheritance of acquired characteristics through the persistence of epigenetic effects through successive generations can also speed up the evolutionary process. These transgenerational environmental influences should spread through a population much more rapidly since it is possible for a large fraction of the population to be subject to the same changes at the same time. There is no need to wait for a single DNA change to spread slowly through a population. The orthodox response to this mechanism is to dismiss it as transient, which it certainly is in some cases. But there are now examples of such transmission over many generations and which show the same degree of strength as standard genetic transmission. Since such effects would not need to occur very frequently, the difficulty in identifying them experimentally would be perfectly understandable. Speciation itself is a rare event.
While there can be considerable uncertainty about the relative contributions of the different mechanisms to evolutionary change, two conclusions seem clear. The first is that, with a variety of mechanisms open to the evolutionary process, the speed of evolution should be faster. The second is that it is probable that the relative contributions varied at different stages in evolution.
The language of neo-darwinism
This page answers the profound, and fundamental, question whether the language of neo-darwinism has been responsible for, and itself expresses, many of the problems with the way in which 20th century biology has been interpreted.
The short answer is ‘yes’. The discourse of neo-darwinism and twentieth century biology in general is intricately linked to the specific philosophical and scientific viewpoints that they represent. It is necessary to deconstruct this discourse since the associated concepts are not required by the scientific discoveries themselves. In fact it can be shown that no biological experiment could possibly distinguish even between completely opposite metaphorical interpretations. The discourse therefore forms an interpretive veneer that can even hide those discoveries in a web of interpretation.
It is also quite difficult to unravel since it is the whole discourse of neo-darwinism that is the problem. Each metaphor reinforces the overall mind-set until it is almost impossible to stand outside it and to appreciate how beguiling it is. Since it has dominated biological science for over half a century, the metaphysical viewpoint represented by this discourse is now so ingrained in the scientific literature that most biological scientists themselves probably don’t recognise its metaphysical nature. Most would probably subscribe to the view that it is merely an accurate description of what experimental work has shown: the discourse in a nutshell is that genes code for proteins that form organisms via a genetic program inherited by subsequent generations and which defines and determines the organism. What is wrong with that? The answer is that almost everything is wrong with it and, sadly, it is not required by the experimental science itself.
The approach used here is, first, to analyse the main metaphors individually, and then to show how they reinforce each other. At the end, I will ask ‘what could be an alternative and better discourse?’
The main problem words are ‘gene’, ‘selfish’, ‘code’, ‘program’, ‘blueprint’, ‘book of life’. We also need to examine secondary concepts like ‘replicator’ and ‘vehicle’.
‘Gene’
Neo-darwinism is a gene-centred theory of evolution. Yet its central entity, the gene, is an unstable concept. Surprising as it may seem, there is no single agreed definition of ‘gene’. Even more seriously, the different definitions have incompatible consequences for the theory.
The word ‘gene’ itself was coined by Johannsen in 1909, but the concept already existed and was based on “the silent assumption [that] was made almost universally that there is a 1:1 relation between genetic factor (gene) and character” (Mayr 1982). Since then, the concept of a gene has changed fundamentally, and this is a major source of confusion when it comes to the question of causation. Its original biological meaning referred to the cause of an inheritable phenotype characteristic, such as /hair/skin colour, body shape and weight, number of legs/arms/wings, to which we could perhaps add more complex traits such as intelligence, personality and sexuality.
The molecular biological definition of a gene is very different. Following the discovery that DNA forms templates for proteins, the definition shifted to locatable regions of DNA sequences with identifiable beginnings and endings. Complexity was added through the discovery of regulatory elements, but the basic cause of phenotype characteristics was still the DNA sequence since that determined which protein was made, which in turn interacted with the rest of the organism to produce the phenotype.
But unless we subscribe to the view that the inheritance of all phenotype characteristics is attributable entirely to DNA sequences (which is just false: DNA never acts outside the context of a complete cell) then genes, as originally conceived, are not the same as the stretches of DNA. According to the original view, genes were necessarily the cause of inheritable phenotypes since that is how they were defined. The issue of causation is now open precisely because the modern definition identifies them instead with DNA sequences.
The original concept of a gene has therefore been taken over and significantly changed by molecular biology. This has undoubtedly led to a great clarification of molecular mechanisms, surely one of the greatest triumphs of twentieth-century biology, and widely acknowledged as such. But the more philosophical consequences of this change for higher level biology are profound and they are much less widely understood.
The difference between the original and the molecular biological definitions of ‘gene’ can be appreciated by noting that most changes in DNA do not necessarily cause a change in phenotype. Organisms are very good at buffering themselves against genomic change. Further analysis of the difference between the definitions can be found in the answer on Slippery Definitions, which also includes an important diagram of the differences.
Some biological scientists have even given up using the word ‘gene’, except in inverted commas. As Beurton et al. (2008) comment “it seems that a cell’s enzymes are capable of actively manipulating DNA to do this or that. A genome consists largely of semi stable genetic elements that may be rearranged or even moved around in the genome thus modifying the information content of DNA.”
The reason that the original and the molecular biological definitions have incompatible consequences is that only the molecular biological definition could be compatible with a strict separation between the ‘replicator’ and the ‘vehicle’. A definition in terms of inheritable phenotypic characteristics (i.e. the original definition) necessarily includes more than the DNA, so that the distinction between replicator and vehicle is blurred.
‘Selfish’
As the lectures show, no possible biological experiment could conceivably distinguish between the selfish gene ‘theory’ and its opposites, such as prisoner or co-operative genes. See the answer What is wrong with The Selfish Gene. This point was conceded long ago by Richard Dawkins at the beginning of his book The Extended Phenotype: ‘I doubt that there is any experiment that could prove my claim’ (Dawkins, 1982, p. 1).
‘Code’
After the discovery of the double helical structure of DNA, it was found that each sequence of three bases in DNA or RNA corresponds to a single amino acid in a protein sequence (see What does DNA do?). These triplet patterns are formed from any combination of the four bases U, C, A and G in RNA and T, C, A and G in DNA. They are often described as the genetic ‘code’, but it is important to understand that this usage of the word ‘code’ is metaphorical and can be confusing.
A real code is an intentional encryption used by humans to communicate. The genetic ‘code’ is not intentional in that sense. The word ‘code’ has unfortunately reinforced the idea that genes are active causes, in much the same way as a computer program directs the computer to obey instructions. The more neutral word ‘template’ would be better. It also expresses the fact that templates are used only when required (activated); they are not themselves active causes. The active causes lie within the cells themselves since they determine the expression patterns for the different cell types and states. These patterns are communicated to the DNA by transcription factors, by methylation patterns and by binding to the tails of histones, all of which influence the pattern and speed of transcription of different parts of the genome. If the word ‘instruction’ is useful at all, it is rather that the cell instructs the genome. As the Nobel-prize winner, Barbara McClintock said, the genome is an ‘organ of the cell’, not the other way round.
Getting the direction of causality in biology wrong is a fundamental mistake with far-reaching consequences. These consequences include the frequent characterisation of genes as the genes ‘for’ this and that.
‘Program’
The idea of ‘le programme génétique’ (‘genetic program’) was first introduced by the French Nobel laureates, Jacques Monod and Francois Jacob. They specifically referred to the way in which early electronic computers were programmed by paper or magnetic tapes: “The programme is a model borrowed from electronic computers. It equates the genetic material with the magnetic tape of a computer” (Jacob 1982). The analogy was that DNA ‘programs’ the cell, tissues and organs of the body just as the code on a computer program determines what the computer does. In principle, the code is independent of the machine that implements it, in the sense that the code itself is sufficient to specify what will happen when the instructions are obeyed. If the program specifies a mathematical computation, for example, it would contain a complete specification of the computation to be performed in the form of complete algorithms. The problem is that no such algorithms can be found in the DNA sequences. What we find is better characterised as a mixture of templates and switches. The ‘templates’ are the triplet sequences that specify the amino acid sequences or the RNA sequences. The ‘switches’ are the locations on the DNA or histones where transcription factors, methylation processes and other controlling processes occur.
Since what we find in the genome sequences are templates and switches, where does the full algorithmic logic of a program lie? Where, for example, do we find the equivalent of ‘IF-THEN-ELSE’ type instructions? The answer is in the cell or organism as a whole, not just in the genome.
Take as an example circadian rhythm. The simplest version of this process depends on a gene that is used as a template for the production of a protein whose concentration then builds up in the cytoplasm. It diffuses through the nuclear membrane and, as the nuclear level increases, it inhibits the transcription process of its own gene. This is a negative feedback loop of the kind that can be represented as implementing a ‘program’ like IF LEVEL X EXCEEDS Y STOP PRODUCING X, BUT IF LEVEL X IS SMALLER THAN Y CONTINUE PRODUCING X. But it is important to note that the implementation of this ‘program’ to produce a 24 hour rhythm depends on rates of protein production by ribosomes, rate of change of concentrations within the cytoplasm, rate of transport across the nuclear membrane, and interaction with the gene transcription control site (the switch). All of this is necessary to produce a feedback circuit that depends on much more than the genome. It depends also on the intricate cellular, tissue and organ structures that are not specified by DNA sequences, which replicate themselves via self-templating, and which are also essential to inheritance across cell and organism generations.
This is true of all such ‘programs’. To call them ‘genetic programs’ is to fuel the misconception that all the logic lies in the one-dimensional DNA sequences. It doesn’t. It also lies in the three dimensional static and dynamic structures of the cells, tissues and organs.
The phrase ‘genetic program’ has therefore encouraged the idea that an organism is fully defined by its genome, whereas in fact it is also defined by its inheritance of cell structure as well as the genome. Moreover, this structure is specific to different species. Cross-species clones do not generally work, and when they do (very rarely) the outcome is determined both by the cytoplasmic structures as well as the DNA – see Immortal genes.
A debate on the motion “No privileged level of causation: An organism is not defined by its genome” was held in Leipzig in July 2012. Image below.

The protagonists were Sydney Brenner and Denis Noble. But the remarkable thing about this debate is that, on the central theme, they were both in agreement with the motion. There was no real debate!
‘Blueprint’
‘Blueprint’ is a variation on the idea of a program. Blueprints, for example as architectural designs for construction of buildings and other structures, existed before the modern idea of a computer program. The word suffers from a similar problem to the concept of a ‘program’, which is that it implies that all the information necessary for the construction of an organism lies in the DNA. This is clearly not true. The complete cell is also required, and its complex structures are inherited by self-templating. The ‘blueprint’, therefore, is the cell as a whole. But that destroys the whole idea of the genome being the full specification.
‘Book of Life’
The genome is often described as the ‘book of life’. This was one of the colourful metaphors used when projecting the idea of sequencing the complete human genome. It was a brilliant public relations move. Who could not be intrigued by reading the ‘book of life’ and unravelling its secrets? And who could resist the promise that, within about a decade, that book would reveal how to treat cancer, heart disease, nervous diseases, diabetes, with a new era of pharmaceutical targets. As we all know, it didn’t happen. Two editorials in 2010 (ten years after the first full draft of the human genome) spelt this out:
“Ten years ago, the first draft of the sequence of the human genome was heralded as the dawn of a new era of genetic medicine………. You might have noticed that it hasn’t [happened]. The medical impact of the human genome project (HGP) has so far been negligible.” (Editorial, Prospect, June 2010).
“The activity of genes is affected by many things not explicitly encoded in the genome, such as how the chromosomal material is packaged up and how it is labelled with chemical markers. Even for diseases like diabetes, which have a clear inherited component, the known genes involved seem to account for only a small proportion of the inheritance…… the failure to anticipate such complexity in the genome must be blamed partly on the cosy fallacies of genetic research. After Francis Crick and James Watson cracked the riddle of DNA’s molecular structure in 1953, geneticists could not resist assuming it was all over bar the shouting. They began to see DNA as the “book of life,” which could be read like an instruction manual. It now seems that the genome might be less like a list of parts and more like the weather system, full of complicated feedbacks and interdependencies.” (Editorial, Nature, 2010).
In response to these editorials I wrote (Noble 2012):
“In 2002 I wrote an article for the magazine of The Physiological Society, Physiology News, explaining why the genome is not the “Book of Life” (Noble, 2002). A reader was so enthused by it that he approached the Editor of Prospect to insist that it deserved a much wider readership and thought that Prospect was the ideal medium. It would have been, but the offer was turned down without question. Probably it didn’t fit the mind set of that time, full of confidence that molecular biology was going, finally, to deliver the goods through exploitation of the genome data.”
“What has changed in the subsequent 8 years, so that even a staff writer for Prospect now expresses the main message of the 2002 article? The answer is that 2010 is the ten year watermark after the sequencing of the genome, when we were promised by the leaders of the Human Genome Project that the benefits for health care would have arrived. Diabetes, hypertension and mental illness were amongst the targets. Science journalists are therefore becoming uneasy that they bought into a promise that has not and, I would argue, could not have been delivered. And they are not alone. The drug industry also bought in, literally so since start-up genomics companies were bought up for hundreds of millions of dollars. The sequencing of genomes has been of great value for basic science, particularly in studies on comparison of genomes for the purposes of evolutionary biology, but the interpretation of the genome data in terms of biological functions (phenotypes) has proved vastly more difficult than anticipated.”
‘The Book of Life’ represents the high watermark of the enthusiasm with which the discourse of neo-darwinism was developed. Its failure speaks volumes: the discourse was not only unnecessary, it was seriously misleading. Yes, there was a good scientific reason for sequencing whole genomes. The benefits to evolutionary biology in particular have been immense. But the discourse promising a peep into the ‘book of life’ and a cure for all diseases was a mistake.
The discourse as a whole
All parts of the discourse of neo-darwinism encourage the use and acceptance of the other parts. Once people have bought in to the idea that the DNA and RNA templates form a ‘code’, the idea of the ‘genetic program’ follows naturally. That leads on to statements like “they [genes] created us body and mind” (The Selfish Gene, 1976). In turn, that leads to the distinction between replicators and vehicles. The mistake lies in accepting the first step, the idea that there is a ‘code’.
The distinction between the replicator and the vehicle can be seen as the culmination of the neo-darwinist discourse. It follows on from accepting the ideas of a code and a program in the genome. If all the algorithms for the logic of life lie in the genome then the rest of the organism does seem to be a disposable vehicle. Only the genome needs to replicate, leaving any old vehicle to carry it.
The distinction however is a linguistic confusion and it is incorrect experimentally. It is a linguistic confusion since the word replicate means to reproduce. Cells also replicate in this sense. They also use templates to do so. The templates are the cell structures themselves. The process is therefore called self-templating and it is just as necessary and just as robust as genome replication. Indeed, faithful genome replication depends on the prior ability of the cell to replicate itself since it is the cell that contains the necessary structures and processes to enable errors in DNA replication to be corrected. See the answer Immortal genes?
The distinction is incorrect experimentally since cross-species cloning shows that cytoplasmic inheritance exists. Another way to make the same point is that the interpretation of the genome depends on the rest of the organism. To use the ‘program’ metaphor, the program is distributed between the genome and the cell.
Notice that the whole discourse is strongly anthropomorphic. This is strange, given that most subscribers to the discourse would wish to avoid anthropomorphising scientific discovery. As Dawkins wrote about the selfish gene metaphor: “I believe it is the literal truth.” When anthropomorphic language becomes confused with ‘literal truth’ we know we are in linguistic trouble.
An alternative discourse

Take some knitting needles and some wool. Knit a rectangle. If you don’t knit, just imagine the rectangle. Or use an old knitted scarf. Now pull on one corner of the rectangle while keeping the opposite corner fixed. What happens? The whole network of knitted knots moves. Now reverse the corners and pull on the other corner. Again the whole network moves. This is a property of networks. Everything ultimately connects to everything else. Any part of the network can be the prime mover, and be the cause of the rest of the network moving and adjusting to the tension.
Now knit a three-dimensional network. Again, imagine it. You probably don’t actually know how to knit such a thing. Pulling on any part of the 3 D structure will cause all other parts to move. It doesn’t matter whether you pull on the bottom, the top or the sides. All can be regarded as equivalent. There is no privileged location within the network.
The 3 D network recalls Waddington’s epigenetic landscape network (also shown here) and is quite a good analogy to biological networks since the third dimension can be viewed as representing the multi-scale nature of biological networks. Properties at the scales of cells, tissues and organs influence activities of elements, such as genes and proteins, at the lower scales. This is sometimes called downward causation, to distinguish it from the reductionist interpretation of causation as upward causation. ‘Down’ and ‘up’ here are also metaphors and should be treated carefully. The essential point is the more neutral statement: there is no privileged level (or scale) of causality. This is necessarily true in organisms which work through many forms of circular causality.

A more complete analysis of this alternative discourse can be found in the article on Biological Relativity, which can also be obtained by downloading the Sourcebook.
The important point about the alternative, relativistic, discourse proposed here is that all the anthropomorphic features of the neo-darwinist discourse can be eliminated, without changing a single biological experimental fact. There may be other discourses that can achieve the same result. It doesn’t really matter which you use. The aim is simply to distance ourselves from the metaphysical baggage that neo-darwinism has brought to biology, made all the worse by the fact that it has been presented as literal truth.
The great physicist, Poincaré, pointed out, in connection with the relativity principle in physics, that the worst philosophical errors are made by those who claim they are not philosophers. They do so because they don’t even recognise the existence of the metaphysical holes they fall into.
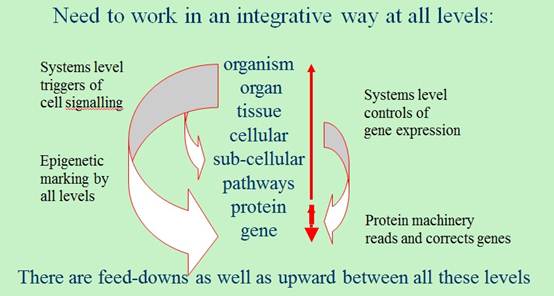
Summary of IUPS 2013 Lecture
This account was posted by a graduate student at the Congress on the same day as the opening ceremony and lecture: 21st July 2013:
http://scienceasadestiny.blogspot.co.uk/2013/07/iups-2013-sir-paul-nurse-and-denis-noble.html
“Physiology moves back onto centre stage: a new synthesis with evolutionary biology”
Denis Noble CBE FRS. About: Eminent researcher in cardio-vascular physiology, current president of the IUPS.
Key point: New insights in physiology are exploding the traditional concepts of Neo-Darwinism evolutionary theory, and opening up a new world of hereditary mechanisms.
“If physiology has moved off centre stage, it is coming back with a vengeance”.
“The genome is an organ of the cell, not a dictator. Control is distributed”.
The focus of this lecture was in demonstrating how the classic views of evolutionary theory are being pulled apart by new physiological advances. According to Neo-Darwinism, evolution is primarily gene-centred and occurs through the gradual accumulation of random mutations. According to the Weismann barrier, the germ line is completely isolated from the parent, hence there is no possibility of acquired traits being inherited. Noble first asked “Are genetic mutations actually random”? Current evidence indicates that genetic mutations follow distinctly non-random patterns throughout the genome. An example of this is P elements, DNA transposons in Drosophila fruit flies – demonstrated to hone in on functionally related areas as they jump between parts of the genome. Noble then explored whether evolution only occurs through gradual assemblies of single mutations. Analysis of the draft human genome sequence in 2001 indicated that the evolution of transcription factors and chromatin binding proteins could not have proceeded one amino acid at a time – rather whole areas and domains must have been shuffled to obtain the current conformation, indicating that mechanisms of reconfiguring the genome must exist. This was illustrated by the example of domestication, a process of introducing changes gradually through generations. However this form of selection “has never led to the formation of a new species. It is a purifying force, not a creative force”. Compare this with hybridisation, which involves mixing up two distinct parental genomes.
Noble also described how our very concept of a gene has changed and the classic linear progression of DNA –> phenotype has been abandoned in favour of a three way interaction between DNA, the environment and the phenotype via a biological network. This explains why knocking out genes rarely reveals their function as the network can compensate for their loss. This was demonstrated effectively by Hillenmeyer et al. who showed that approximately 80 % of knock out gene mutations in yeast are silent unless additional environmental constraints are imposed.
Noble then moved on to ask “Why should a physiologist be concerned with evolutionary biology?”. Traditional evolutionary views are gene centred, yet physiological research is demonstrating that organisms can “immune themselves from the genome”. Furthermore, information transmission is not a one-way process as organisms can impose downward control onto DNA through cell signalling, transcription factors and epigenetic modification. An example of this is provided by work on rats showing that regular grooming in early life makes the mature adult less fearful – is grooming time limited in colonies stressed by predation or starvation? Another exciting illustration is the production of cross species fish by placing a carp nucleus into an enucleated cell from a goldfish. In the rare circumstances when this produces an embryo, the skeletal configuration is intermediate between the two, but much more similar to the goldfish. Hence, information cannot be transmitted solely by the DNA but must be influenced by maternal factors in the egg cytoplasm. Work on the nematode worm C. elegans meanwhile, has revealed that epigenetic changes can be incredibly robust. Here the inheritance of antiviral RNA molecules was demonstrated for up to 100 generations, even though the DNA template had been lost; inheritance had been achieved through RNA polymerase amplification in the cytoplasm. Our view of the DNA machinery should echo that of Barbara McClintock, who viewed DNA as a highly sensitive organ that can detect and respond to unexpected events. Noble cited how genome reorganisation may also occur through the lateral acquisition of new DNA material from unrelated cells, such as the ingestion of the prokaryote cells which became reduced to mitochondria and chloroplasts. The central concept of this stimulating lecture was clear: the genome is NOT isolated from the environment and furthermore, acquired characteristics can be inherited. Perhaps Lamarck wasn’t so wrong after all.
How widespread is trans-generational inheritance of acquired phenotype characteristics?
Since this issue is central and requires careful attention to the details of the experimental evidence, I have included abstracts from the articles cited. This form of inheritance is now firmly established.
“Many maternal effects have subsequently been observed, and non-genomic transmission of disease risk has been firmly established (P. Gluckman & Hanson, 2004; P. D. Gluckman, Hanson, & Beedle, 2007). A study done in Scandinavia clearly shows the transgenerational effect of food availability to human grandparents influencing the longevity of grandchildren (Kaati, Bygren, Pembrey, & Sjostrom, 2007; Pembrey et al., 2006).”
Gluckman, P., & Hanson, M. (2004). The Fetal Matrix. Evolution, Development and Disease. Cambridge: Cambridge University Press.
Gluckman, P. D., Hanson, M. A., & Beedle, A. S. (2007). Non-genomic transgenerational inheritance of disease risk. Bioessays, 29, 145-154. http://www.ncbi.nlm.nih.gov/pubmed/17226802
That there is a heritable or familial component of susceptibility to chronic non-communicable diseases such as type 2 diabetes, obesity and cardiovascular disease is well established, but there is increasing evidence that some elements of such heritability are transmitted non-genomically and that the processes whereby environmental influences act during early development to shape disease risk in later life can have effects beyond a single generation. Such heritability may operate through epigenetic mechanisms involving regulation of either imprinted or non-imprinted genes but also through broader mechanisms related to parental physiology or behaviour. We review evidence and potential mechanisms for non-genomic transgenerational inheritance of ‘lifestyle’ disease and propose that the ‘developmental origins of disease’ phenomenon is a maladaptive consequence of an ancestral mechanism of developmental plasticity that may have had adaptive value in the evolution of generalist species such as Homo sapiens.
Kaati, G., Bygren, L. O., Pembrey, M. E., & Sjostrom, M. (2007). Transgenerational response to nutrition, early life circumstances and longevity European Journal of Human Genetics, 15, 784-790. http://www.ncbi.nlm.nih.gov/pubmed/17457370
Nutrition might induce, at some loci, epigenetic or other changes that could be transmitted to the next generation impacting on health. The slow growth period (SGP) before the prepubertal peak in growth velocity has emerged as a sensitive period where different food availability is followed by different transgenerational response (TGR). The aim of this study is to investigate to what extent the probands own childhood circumstances are in fact the determinants of the findings. In the analysis, data from three random samples, comprising 271 probands and their 1626 parents and grandparents, left after exclusions because of missing data, were utilized. The availability of food during any given year was classified based on regional statistics. The ancestors’ SGP was set at the ages of 8-12 years and the availability of food during these years classified as good, intermediate or poor. The probands’ childhood circumstances were defined by the father’s ownership of land, the number of siblings and order in the sibship, the death of parents and the parents’ level of literacy. An earlier finding of a sex-specific influence from the ancestors’ nutrition during the SGP, going from the paternal grandmother to the female proband and from the paternal grandfather to the male proband, was confirmed. In addition, a response from father to son emerged when childhood social circumstances of the son were accounted for. Early social circumstances influenced longevity for the male proband. TGRs to ancestors’ nutrition prevailed as the main influence on longevity.
Pembrey, M. E., Bygren, L. O., Kaati, G., Edvinsson, S., Northstone, K., Sjostrom, M., . . . ALSPAC_study_team. (2006). Sex-specific, male-line transgenerational responses in humans. European Journal of Human Genetics, 14, 159-166. http://www.ncbi.nlm.nih.gov/pubmed/16391557
Transgenerational effects of maternal nutrition or other environmental ‘exposures’ are well recognised, but the possibility of exposure in the male influencing development and health in the next generation(s) is rarely considered. However, historical associations of longevity with paternal ancestors’ food supply in the slow growth period (SGP) in mid childhood have been reported. Using the Avon Longitudinal Study of Parents and Children (ALSPAC), we identified 166 fathers who reported starting smoking before age 11 years and compared the growth of their offspring with those with a later paternal onset of smoking, after correcting for confounders. We analysed food supply effects on offspring and grandchild mortality risk ratios (RR) using 303 probands and their 1818 parents and grandparents from the 1890, 1905 and 1920 Overkalix cohorts, northern Sweden. After appropriate adjustment, early paternal smoking is associated with greater body mass index (BMI) at 9 years in sons, but not daughters. Sex-specific effects were also shown in the Overkalix data; paternal grandfather’s food supply was only linked to the mortality RR of grandsons, while paternal grandmother’s food supply was only associated with the granddaughters’ mortality RR. These transgenerational effects were observed with exposure during the SGP (both grandparents) or fetal/infant life (grandmothers) but not during either grandparent’s puberty. We conclude that sex-specific, male-line transgenerational responses exist in humans and hypothesise that these transmissions are mediated by the sex chromosomes, X and Y. Such responses add an entirely new dimension to the study of gene-environment interactions in development and health.
Contrary to a widespread view that such effects always die out quickly, this form of inheritance can be just as strong as conventional genetic inheritance.
“Their article (Nelson et al, 2012) begins by noting that many environmental agents and genetic variants can induce heritable epigenetic changes that affect phenotypic variation and disease risk in many species. Moreover, these effects persist for many generations and are as strong as conventional genetic inheritance (Cuzin & Rassoulzadegan, 2010; Guerrero-Bosagna & Skinner, 2012; Jirtle & Skinner, 2007; Nelson & Nadeau, 2010; Richards, 2006; Youngson & Whitelaw, 2008).”
Cuzin, F., Grandjean, V., & Rassoulzadegan, M. (2008). Inherited variation at the epigenetic level: paramutation from the plant to the mouse. Curr Opin Genet Dev, 18(2), 193-196. http://www.ncbi.nlm.nih.gov/pubmed/18280137
In contrast with a wide definition of the ‘epigenetic variation’, including all changes in gene expression that do not result from the alteration of the gene structure, a more restricted class had been defined, initially in plants, under the name ‘paramutation’. It corresponds to epigenetic modifications distinct from the regulatory interactions of the cell differentiation pathways, mitotically stable and sexually transmitted with non-Mendelian patterns. This class of epigenetic changes appeared for some time restricted to the plant world, but examples progressively accumulated of epigenetic inheritance in organisms ranging from mice to humans. Occurrence of paramutation in the mouse and possible mechanisms were then established in the paradigmatic case of a mutant phenotype maintained and hereditarily transmitted by wild-type homozygotes. Together with the recent findings in plants indicative of a necessary step of RNA amplification in the reference maize paramutation, the mouse studies point to a new role of RNA, as an inducer and hereditary determinant of epigenetic variation. Given the known presence of a wide range of RNAs in human spermatozoa, as well as a number of unexplained cases of familial disease predisposition and transgenerational maintenance, speculations can be extended to possible roles of RNA-mediated inheritance in human biology and pathology
Guerrero-Bosagna, C., & Skinner, M. K. (2012). Environmentally-induced epigenetic transgenerational inheritance of phenotype and disease Molecular and cellular endocrinology, 354, 3-8. http://www.ncbi.nlm.nih.gov/pubmed/22020198
Environmental epigenetics has an important role in regulating phenotype formation or disease etiology. The ability of environmental factors and exposures early in life to alter somatic cell epigenomes and subsequent development is a critical factor in how environment affects biology. Environmental epigenetics provides a molecular mechanism to explain long term effects of environment on the development of altered phenotypes and “emergent” properties, which the “genetic determinism” paradigm cannot. When environmental factors permanently alter the germ line epigenome, then epigenetic transgenerational inheritance of these environmentally altered phenotypes and diseases can occur. This environmental epigenetic transgenerational inheritance of phenotype and disease is reviewed with a systems biology perspective.
Jirtle, R. L., & Skinner, M. K. (2007). Environmental epigenomics and disease susceptibility Nature Reviews Genetics, 8, 253-262. http://www.ncbi.nlm.nih.gov/pubmed/17363974
Epidemiological evidence increasingly suggests that environmental exposures early in development have a role in susceptibility to disease in later life. In addition, some of these environmental effects seem to be passed on through subsequent generations. Epigenetic modifications provide a plausible link between the environment and alterations in gene expression that might lead to disease phenotypes. An increasing body of evidence from animal studies supports the role of environmental epigenetics in disease susceptibility. Furthermore, recent studies have demonstrated for the first time that heritable environmentally induced epigenetic modifications underlie reversible transgenerational alterations in phenotype. Methods are now becoming available to investigate the relevance of these phenomena to human disease
Nelson VR, Heaney JD, Tesar PJ, Davidson NO & Nadeau JH (2012). Transgenerational epigenetic effects ofApobec1 deficiency on testicular germ cell tumor susceptibility and embryonic viability. Proc Natl Acad Sci U S A 109, E2766–E2773 http://www.ncbi.nlm.nih.gov/pubmed/22923694
Environmental agents and genetic variants can induce heritable epigenetic changes that affect phenotypic variation and disease risk in many species. These transgenerational effects challenge conventional understanding about the modes and mechanisms of inheritance, but their molecular basis is poorly understood. The Deadend1 (Dnd1) gene enhances susceptibility to testicular germ cell tumors (TGCTs) in mice, in part by interacting epigenetically with other TGCT modifier genes in previous generations. Sequence homology to A1cf, the RNA-binding subunit of the ApoB editing complex, raises the possibility that the function of Dnd1 is related to Apobec1 activity as a cytidine deaminase. We conducted a series of experiments with a genetically engineered deficiency of Apobec1 on the TGCT-susceptible 129/Sv inbred background to determine whether dosage of Apobec1 modifies susceptibility, either alone or in combination with Dnd1, and either in a conventional or a transgenerational manner. In the paternal germ-lineage, Apobec1 deficiency significantly increased susceptibility among heterozygous but not wild-type male offspring, without subsequent transgenerational effects, showing that increased TGCT risk resulting from partial loss of Apobec1 function is inherited in a conventional manner. By contrast, partial deficiency in the maternal germ-lineage led to suppression of TGCTs in both partially and fully deficient males and significantly reduced TGCT risk in a transgenerational manner among wild-type offspring. These heritable epigenetic changes persisted for multiple generations and were fully reversed after consecutive crosses through the alternative germ-lineage. These results suggest that Apobec1 plays a central role in controlling TGCT susceptibility in both a conventional and a transgenerational manner.
Nelson, V. R., & Nadeau, J. H. (2010). Transgenerational genetic effects. Epigenomics, 2, 797-806. http://www.ncbi.nlm.nih.gov/pubmed/22122083
Since Mendel, studies of phenotypic variation and disease risk have emphasized associations between genotype and phenotype among affected individuals in families and populations. Although this paradigm has led to important insights into the molecular basis for many traits and diseases, most of the genetic variants that control the inheritance of these conditions continue to elude detection. Recent studies suggest an alternative mode of inheritance where genetic variants that are present in one generation affect phenotypes in subsequent generations, thereby decoupling the conventional relations between genotype and phenotype, and perhaps, contributing to ‘missing heritability’. Under some conditions, these transgenerational genetic effects can be as frequent and strong as conventional inheritance, and can persist for multiple generations. Growing evidence suggests that RNA mediates these heritable epigenetic changes. The primary challenge now is to identify the molecular basis for these effects, characterize mechanisms and determine whether transgenerational genetic effects occur in humans.
Richards, E. J. (2006). Inherited epigenetic variation – revisiting soft inheritance Nature Reviews Genetics, 7, 395-401. http://www.ncbi.nlm.nih.gov/pubmed/16534512
Phenotypic variation is traditionally parsed into components that are directed by genetic and environmental variation. The line between these two components is blurred by inherited epigenetic variation, which is potentially sensitive to environmental inputs. Chromatin and DNA methylation-based mechanisms mediate a semi-independent epigenetic inheritance system at the interface between genetic control and the environment. Should the existence of inherited epigenetic variation alter our thinking about evolutionary change?
Youngson, N. A., & Whitelaw, E. (2008). Transgenerational epigenetic effects Annual Review of Genomics and Human Genetics, 9, 233-257. http://www.ncbi.nlm.nih.gov/pubmed/18767965
Transgenerational epigenetic effects include all processes that have evolved to achieve the nongenetic determination of phenotype. There has been a long-standing interest in this area from evolutionary biologists, who refer to it as non-Mendelian inheritance. Transgenerational epigenetic effects include both the physiological and behavioral (intellectual) transfer of information across generations. Although in most cases the underlying molecular mechanisms are not understood, modifications of the chromosomes that pass to the next generation through gametes are sometimes involved, which is called transgenerational epigenetic inheritance. There is a trend for those outside the field of molecular biology to assume that most cases of transgenerational epigenetic effects are the result of transgenerational epigenetic inheritance, in part because of a misunderstanding of the terms. Unfortunately, this is likely to be far from the truth.
RNA transmitted changes independent of DNA have been followed in planarians for 100 generations:
Rechavi O, Minevish G & Hobert O (2011). Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans. Cell 147, 1248–1256. http://www.cell.com/retrieve/pii/S0092867411013419#Summary
Induced expression of the Flock House virus in the soma of C. elegans results in the RNAi-dependent production of virus-derived, small-interfering RNAs (viRNAs), which in turn silence the viral genome. We show here that the viRNA-mediated viral silencing effect is transmitted in a non-Mendelian manner to many ensuing generations. We show that the viral silencing agents, viRNAs, are transgenerationally transmitted in a template-independent manner and work in trans to silence viral genomes present in animals that are deficient in producing their own viRNAs. These results provide evidence for the transgenerational inheritance of an acquired trait, induced by the exposure of animals to a specific, biologically relevant physiological challenge. The ability to inherit such extragenic information may provide adaptive benefits to an animal.
Transgenerational epigenetic effects in the brain are reviewed in
Bohacek J, Gapp K, Saab BJ & Mansuy IM. (2013). Transgenerational Epigenetic Effects on Brain Functions. Biological Psychiatry 73, 313-320.
http://www.biologicalpsychiatryjournal.com/article/S0006-3223(12)00729-9/abstract
Psychiatric diseases are multifaceted disorders with complex etiology, recognized to have strong heritable components. Despite intense research efforts, genetic loci that substantially account for disease heritability have not yet been identified. Over the last several years, epigenetic processes have emerged as important factors for many brain diseases, and the discovery of epigenetic processes in germ cells has raised the possibility that they may contribute to disease heritability and disease risk. This review examines epigenetic mechanisms in complex diseases and summarizes the most illustrative examples of transgenerational epigenetic inheritance in mammals and their relevance for brain function. Environmental factors that can affect molecular processes and behavior in exposed individuals and their offspring, and their potential epigenetic underpinnings, are described. Possible routes and mechanisms of transgenerational transmission are proposed, and the major questions and challenges raised by this emerging field of research are considered.
Transgenerational epigenetic effects underly the inheritance of sensitivity to odors in mice:
Dias BG & Ressler KJ. (2013). Parental olfactory experience influences behavior and neural structure in subsequent generations. Nature Neuroscience, doi:10.1038/nn.3594
Using olfactory molecular specificity, we examined the inheritance of parental traumatic exposure, a phenomenon that has been frequently observed, but not understood. We subjected F0 mice to odor fear conditioning before conception and found that subsequently conceived F1 and F2 generations had an increased behavioral sensitivity to the F0-conditioned odor, but not to other odors. When an odor (acetophenone) that activates a known odorant receptor (Olfr151) was used to condition F0 mice, the behavioral sensitivity of the F1 and F2 generations to acetophenone was complemented by an enhanced neuroanatomical representation of the Olfr151 pathway. Bisulfite sequencing of sperm DNA from conditioned F0 males and F1 naive offspring revealed CpG hypomethylation in the Olfr151 gene. In addition, in vitro fertilization, F2 inheritance and cross-fostering revealed that these transgenerational effects are inherited via parental gametes. Our findings provide a framework for addressing how environmental information may be inherited transgenerationally at behavioral, neuroanatomical and epigenetic levels.
For examples of epigenetic inheritance in plants see
Pennisi, E (2013) Evolution Heresy? Epigenetics Underlies Heritable Plant Traits Science 341 6 September http://www.sciencemag.org/content/341/6150/1055.summary
“For some evolutionary biologists, just hearing the term epigenetics raises hackles. They balk at suggestions that something other than changes in DNA sequences—such as the chemical addition of methyl groups to DNA or other so-called epigenetic modifications— has a role in evolution. All of which guarantees that a provocative study presented at an evolutionary biology meeting …. last month will get close scrutiny. It found that heritable changes in plant flowering time and other traits were the result of epigenetics alone, unaided by any sequence changes.”
Colomé-Tatché M, Cortijo S, Wardenaar R, Morgado L, Lahouze B, Sarazin A, Etcheverry M, Martin A, Feng S, Duvernois-Berthet E, Labadie K, Wincker P, Jacobsen SE, Jansen RC, Colot V, Johannes F (2012). Features of the Arabidopsis recombination landscape resulting from the combined loss of sequence variation and DNA methylation. Proc. Natl. Acad. Sci. USA doi:10.1073/pnas.1212955109. Research reported by Frank Johannes (Groningen) http://www.johanneslab.org/
Schmitz, R.J. et al (2011) Transgenerational Epigenetic Instability is a source of Novel Methylation Variants. Science, 334, 369-373. “We examined spontaneously occurring variation in DNA methylation in Arabidopsis thaliana plants propagated by single-seed descent for 30 generations…… transgenerational epigenetic variation in DNA methylation may generate new allelic states that alter transcription, providing a mechanism for phenotypic diversity in the absence of genetic mutation.” “Regardless of their origin, the majority of epialleles identified in this study are meiotically stable and heritable across many generations in this population.”
http://www.sciencemag.org/content/334/6054/369.abstract
Schmitz, R.J. et al (2013) Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome Research, 23, 1663-1674. “a comprehensive study of the patterns and heritability of methylation variants in a complex genetic population over multiple generations, paving the way for understanding how methylation variants contribute to phenotypic variation.”
http://genome.cshlp.org/content/early/2013/08/29/gr.152538.112.full.pdf+html
The big question now is how large a role these forms of inheritance have played in the evolutionary process. But that is a question that applies to all the proposed mechanisms of evolutionary change and also to the ways in which they must have interacted. Articles relevant to that question include:
Hua, Z. (2013) Epigenomic programming contributes to the genomic drift evolution of the F-Box protein superfamily in Arabidopsis. PNAS, 110, 16927–16932. “Comparisons within expanding sequence databases have revealed a dynamic interplay among genomic and epigenomic forces in driving plant evolution. Such forces are especially obvious within the F-Box (FBX) superfamily, one of the largest and most polymorphic gene families in land plants, where its frequent lineage-specific expansions and contractions provide an excellent model to assess how genetic variation impacted gene function before and after speciation.” “…reversible epigenomic modifications helped shape FBX gene evolution by transcriptionally suppressing the adverse effects of gene dosage imbalance and harmful FBX alleles that arise during genomic drift, while simultaneously allowing innovations to emerge through epigenomic reprogramming.”
http://www.ncbi.nlm.nih.gov/pubmed/24082131
Takuno, S & Gaut B.S. (2013) Gene body methylation is conserved between plant orthologs and is of evolutionary consequence. PNAS, 110, 1797-1802. “Gene body methylation was strongly conserved between orthologs of the two species and affected a biased subset of long, slowly evolving genes. Because gene body methylation is conserved over evolutionary time, it shapes important features of plant genome evolution, such as the bimodality of G+C content among grass genes.”
http://www.ncbi.nlm.nih.gov/pubmed/23319627
The following article is a useful critique of the inheritability of stress-induced chromatin changes in plants, and lays out some criteria to be used in further work.
Pecinka, A. & Scheid, O.M. (2012) Stress-Induced Chromatin Changes: A Critical View on Their Heritability. Plant Cell Physiol 53, 801-808. “We propose a set of criteria that should be applied to substantiate the data for stress-induced, chromatin-encoded new traits. Well-controlled stress treatments, thorough phenotyping and application of refined genome-wide epigenetic analysis tools should be helpful in moving from interesting observations towards robust evidence.” “plants are good candidates for a further, unprepossessed search. Constant refinement of chromatin analysis tools and growing genetic information, also for non-model species, together with the criteria listed here, will help answer whether it is time for a renaissance of Lamarck’s ideas.”
http://pcp.oxfordjournals.org/content/53/5/801.full
This is also a valuable review:
O’Malley R.C. & Ecker, J.R. (2012) Epiallelic Variation in Arabidopsis thaliana. Cold Spring Harbour Symp Quant Biol. 77, 135-145. “Genotype is the primary determinate of phenotype. During the past two decades, however, there has been an emergent recognition of the epigenotype, a separate layer of heredity distinct from the primary DNA sequence that can have profound effects on phenotype.” “We discuss examples of epialleles that have been created in the laboratory and others that have been identified in natural populations, because these two models provide complementary information regarding the genetic pathways, mechanisms of transmission, and biological and evolutionary context for the role of the epigenotype in phenotypic variation.”
Is the proposed Integrative Synthesis really new?
The article proposes a new synthesis which differs in four major respects from the traditional (modern) synthesis:
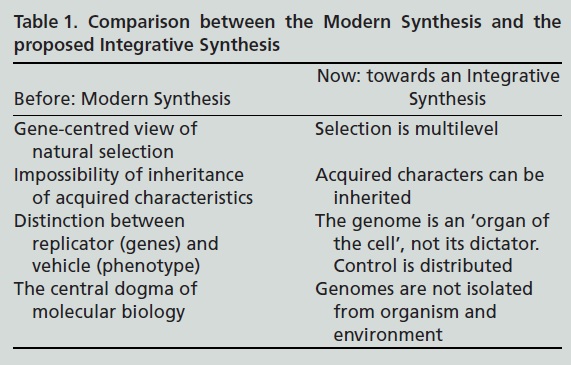
But is this really a new proposal? Havn’t we heard about the demise of neo-Darwinism too often?
“The full extent of this feedback from function to inheritance remains to be assessed, but it cannot be doubted that it runs counter to the spirit of the Modern Synthesis. The challenge now is how to construct a new Synthesis to take account of this development. In Table 1, I call this the Integrative Synthesis. I believe that in the future, the Modern Synthesis and the elegant mathematics that it gave rise to, for example in the various forms and developments of the Price equation, will be seen as only one of the processes involved, a special case in certain circumstances, just as Newtonian mechanics remains as a special case in the theory of relativity.”
The problems with the modern synthesis are that it is restrictive and that it is dogmatic.
“What went wrong in the mid-20th century that led us astray for so long? The answer is that all the way from the Weismann barrier experiments in 1893 (which were very crude experiments indeed) through to the formulation of the central dogma of molecular biology in 1970, too much was claimed for the relevant experimental results, and it was claimed too dogmatically. Demonstrating, as Weismann did, that cutting the tails off many generations of mice does not result in tail-less mice shows, indeed, that this particular induced characteristic is not inherited, but it obviously could not exclude other mechanisms. The mechanisms found recently are far more subtle. Likewise, the demonstration that protein sequences do not form a template for DNA sequences should never have been interpreted to mean that information cannot pass from the organism to its genome. Barbara McClintock deservedly gets the last laugh; the genome is indeed an ‘organ of the cell’.”
The idea of a more nuanced multi-mechanism synthesis is indeed not new. I am delighted, for example, to acknowledge the similarity of my ideas to those of Eugene Koonin (2009), Messoudi et al. (2013) and Laland et al (2013). The problem is that the dogmatism of neo-Darwinism has prevented open admission of the change. The change has already happened in the minds of those who listen to the experimental evidence.
Eugene Koonin, The Origin at 150: Is a new evolutionary synthesis in sight? Trends in Genetics, 25, November 2009, pp. 473-475. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2784144/
Eugene Koonin, Darwinian evolution in the light of genomics, Nucleic Acids Research, 37, 2009, pp. 1011-1034. http://www.ncbi.nlm.nih.gov/pubmed/19213802
Mesoudi A, Blanchet S, Charmentier A, Danchin E, Fogarty L, Jablonka E, Laland KN, Morgan TJH, Mueller GB, Odling-Smee FJ & Pojol B. (2013). Is non-genetic inheritance just a proximate mechanism? A corroboration of the extended evolutionary synthesis. Biological Theory 7, 189–195. http://link.springer.com/article/10.1007/s13752-013-0091-5
Laland, K. N., Odling-Smee, F. J., Hoppitt, W., and Uller, T. (2013) More on how and why: a response to commentaries, Biology and Philosophy DOI: 10.1007/s10539-013-9380-4.
Quotes from Koonin that resemble my text:
“In the post-genomic era, all the major tenets of the modern synthesis have been, if not outright overturned, replaced by a new and incomparably more complex vision of the key aspects of evolution.”
“The discovery of pervasive HGT [horizontal gene transfer] and the overall dynamics of the genetic universe destroys not only the tree of life as we knew it but also another central tenet of the modern synthesis inherited from Darwin, namely gradualism. In a world dominated by HGT, gene duplication, gene loss and such momentous events as endosymbiosis, the idea of evolution being driven primarily by infinitesimal heritable changes in the Darwinian tradition has become untenable.”
“The edifice of the modern synthesis has crumbled, apparently, beyond repair”.
“The exclusive focus of Modern Synthesis on natural selection acting on random genetic variation has been replaced with a plurality of complementary, fundamental evolutionary processes and patterns.”
Laland et al (2013) also express many of the same ideas, and it is particularly important to note that they have also pinpointed the concept of causation as one of the problems:
“We (like many other developmentally minded evolutionists, e.g. West-Eberhard 2003) believe that resistance to these ideas derives in part from implicit models of causation that can channel thinking on these topics, leading to the neglect of potentially important explanations. For instance, in their recent review of phenotypic plasticity’s impacts on speciation, where extensive evidence that plasticity is evolutionarily consequential was presented, Pfennig et al. (2010, p. 459) nonetheless conclude that “recent reviews of speciation generally fail to discuss phenotypic plasticity, indicating that workers in this field do not recognize a significant role for plasticity in speciation”.”
which may be compared to my article:
“A central feature of the Integrative Synthesis is a radical revision of the concept of causality in biology. A priori there is no privileged level of causation. This is the principle that I have called the theory of biological relativity (Noble, 2008, 2012)…… Control is therefore distributed, some of which is inherited independently of DNA sequences. The revision of the concept will also recognize the different forms of causality. DNA sequences are best viewed as passive causes, because they are used only when the relevant sequences are activated. DNA on its own does nothing. The active causes lie within the control networks of the cells, tissues and organs of the body.”
Laland et al also compare their position with the Modern Synthesis in the form of a table:
The compatibility of this table with my own is obvious. I do not claim any priority in expressing my ideas and I am delighted to discover others who see the evidence in much the same way. Where I have gone a little bit further is in pointing out how physiology comes back onto centre stage as the study of function: “the organism should never have been relegated to the role of mere carrier of its genes.”

What’s wrong with The Selfish Gene?
The Selfish Gene by Richard Dawkins is a highly persuasive presentation of the neo-darwinist gene-centred view of evolution. I have analysed it in detail in an article published in 2011:
Noble D, (2011) Neo-Darwinism, the Modern Synthesis and selfish genes: are they of use in physiology? Journal of Physiology, 589, 1007-1015.
Available here: http://musicoflife.website/pdfs/Selfish%20Genes.pdf
As with all of Dawkins’ books, the writing is impressive. The Selfish Gene is colourful, convincing and unforgettable – until one tries to analyse it by the standard philosophical and scientific criteria. Then it unravels.
First, it unravels because there has been confusion over whether the title was or was not meant to be metaphorical. It is clear that when it was first written in 1976, and even as late as 1981, Dawkins’ position was “that was no metaphor. I believe it is the literal truth”.
Dawkins R (1981). In defence of selfish genes. Philosophy 56, 556–573 http://www.jstor.org/discover/10.2307/3750888?uid=3738032&uid=2&uid=4&sid=21102396324337
I strongly believe that was indeed the case. It is hard to read The Selfish Gene without supposing that its author was intent on conveying hard and incontestable scientific truth. The book exudes that kind of confidence in its message. So, why couldn’t it be ‘the literal truth’? The reason is provided by Dawkins himself in his 1981 article. He tries to explain why it is literal truth by explaining that it is a metaphor! The sentence I quoted above continues “provided certain key words are defined in the particular ways favoured by biologists”. A metaphor is precisely ‘certain key words defined in particular ways’. The whole point of a metaphor is that the meaning of the word is changed from its normal literal meaning.
This confusion is symptomatic of a deeper confusion. As pure metaphor, the idea suffers from the difficulty that no experiment could ever distinguish between the selfish gene view and opposing metaphors, such as co-operative or prisoner genes. In my 2011 article I attempt to find a way out of this problem by asking whether there are ways of unpacking the metaphor to give the idea some empirical leverage, so that it could at least become testable. One way of doing that is to conflate the terms “selfish” and “successful”. A selfish bit of DNA is then the one that succeeds in increasing its frequency in the gene pool. But what this does is to make the hypothesis empty, circular. The only prediction of the hypothesis, i. e. success in increasing frequency in the gene pool, is also the definition of the hypothesis’ central entity. As I say in my 2011 article “It is a strange hypothesis that uses its own definition of its postulated entity as its only prediction.” Joan Roughgarden has also spotted that conflating “selfish” and “successful” makes the hypothesis empty: “But that vacates the meaning of selfish. “Selfish gene” and “successful gene” are not the same thing.”
Roughgarden, JE (2009) The Genial Gene. Deconstructing Darwinian Selfishness. Berkeley, University of California Press
We have to live therefore with the uncomfortable fact that it “is clearly metaphorical metaphysics, and rather poor metaphysics at that since, as we have seen, it is essentially empty as a scientific hypothesis, at least in physiological science” (Noble 2011).
Slippery definitions: how neo-darwinism accommodates contrary findings
There is a long history to this. There have been many points at which neo-darwinism has been challenged experimentally. The reactions can be characterised as a mixture of assimilation and denial.
An early example of assimilation is the reaction to the work of Conrad Waddington who first coined the term ‘epigenetics’. Waddington showed that an acquired characteristic in fruit flies could be assimilated after a number of generations of selection for the trait. After that it became inheritable in the standard way without the environmental stimulus that caused the character to be acquired. In the Experimental Physiology article I write
The Modern Synthesists should not have dismissed Waddington’s experiments, for example, as simply ‘a special case of the evolution of phenotypic plasticity’ (Arthur, 2010). Of course, the Modern Synthesis can account for the inheritance of the potential for plasticity, but what it cannot allow is the inheritance of a specific acquired form of that plasticity. Waddington’s experiments demonstrate precisely inheritance of specific forms of acquired characteristics, as he claimed himself in the title of his paper (Waddington, 1942). After all, the pattern of the genome is as much inherited as its individual components, and those patterns can be determined by the environment.
Notice that this also illustrates how neo-darwinism tends to be slippery when it defines a gene. Defining genes as individual protein-coding sequences ignores inheritance of characteristics that depend on the pattern of the genome. Waddington’s experiments show that it is perfectly plausible that a characteristic first arises through environmentally-induced change and then becomes ‘locked in’ to the genome pattern so that it becomes standard genomic inheritance. No mutations would be required and the characteristic could appear in a substantial fraction of the population rather than a single individual. We don’t know how often this process may have occurred during evolution. I suspect that many of the additional mechanisms of variation may become ‘locked in’ in this way.
A ‘gene’ in this sense can be a new combination of alleles that allows a new phenotype to develop. The mistake is to ‘atomise’ the concept of a gene. That is another important difference between the original definition in terms of phenotypes and the molecular biological definition in terms of ‘atomised’ DNA elements.
An example of what I would call denial is provided by an exchange during the 2009 Oxford debate that I chaired between Margulis and Dawkins. The transcript includes Dawkins’ reaction to the example of cellular inheritance independent of the genome in Paramecium:
“One example you might have meant is Sonneborn’s Paramecium where you cut a bit of the pellicle [ciliate cortex] and twist it ‘round. Well, if that’s true, and is indeed a non-DNA form of heredity, that’s absolutely fine. I would embrace that gladly as a new “honorary” gene. That’s fine. [Groans from the audience] Why not, why not?” (transcript, page 21)
The groans from the audience surely indicate something slippery here. I’d call it ‘having it both ways’. The whole point of the discussion was whether inheritance is determined by genes alone. Sure, we can go on redefining ‘gene’ until we have exhausted all the possibilities. But a statement that says everything also says nothing. The denial here is a denial of a counter-example to neo-darwinism by redefining what is meant by a gene. As I explain in the lectures the central confusion lies precisely in the slippage between defining genes as DNA sequences and their original definition in terms of inheritable phenotypes. Defined as the latter, there can, by definition, be no exceptions. Defined as the former, there clearly are exceptions.
The relevant slide in the lecture is:
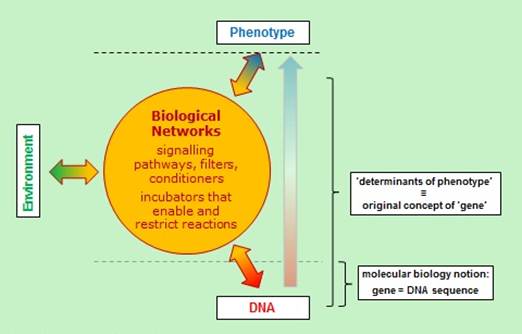
Relations between genes, environment and phenotype characters according to current physiological and biochemical understanding This diagram represents the interaction between genes (DNA sequences), environment and phenotype as occurring through biological networks. The causation occurs in both directions between all three influences on the networks. This view is very different from the idea that genes ‘cause’ the phenotype (right hand arrow). This diagram also helps to explain the difference between the original concept of a gene as the cause of a particular phenotype and the modern definition as a DNA sequence. For further description and analysis of the ideas behind this diagram see Kohl et al. (2010) Clinical Pharmacology and Therapeutics 88, 25–33
The sense in which neo-darwinism has been falsified in this case is precisely that epigenetic inheritance shows that characteristics of the biological networks can be inherited independently of DNA. Examples of that are now rapidly accumulating.
Are these kinds of confusions still occurring? A recent example is provided by
Zuk et al (2012) The mystery of missing heritability: Genetic interactions create phantom heritability. PNAS, 109, 1193-1198.
which contains an equivalent to Dawkins’ invention of ‘honorary gene’ by inventing the term ‘phantom heritability’. In this case, the interactions that necessarily include the biological networks are apparently assimilated into standard genetics (hence the term ‘genetic interactions’), when in fact there must be much more inherited than the relevant DNA sequences. The authors write “In short, missing heritability need not directly correspond to missing variants, because current estimates of total heritability may be significantly inflated by genetic interactions.” The matter should not be closed off in this way since the question whether this does or does not involve inheritance of epigenetic effects must remain open to further experimentation. And it would be better to recognise this by avoiding terms like ‘genetic interactions’ that leave it unclear what precisely is involved but which also prejudice the conclusion in favour of standard genetic inheritance.
Redefinition of the central entity in neo-darwinism, i.e. the ‘gene’, leads to unintended consequences. Admitting ‘honorary’ or ‘phantom’ genes that refer entirely or partly to cytoplasmic inheritance makes nonsense of the distinction between ‘replicator’ and ‘vehicle’. The ‘vehicle’ becomes part of the ‘replicator’.
It is anyway! See Immortal genes.
New Synthesis or Extended Synthesis?
Even some of those who accept that we need to move on from the Modern Synthesis think that it would be simpler just to extend it by incorporating new experimental observations and the mechanisms they identify. So, would an Extended (Modern) Synthesis be sufficient? To the Modern Synthesis, we could add all the other mechanisms (inheritance of acquired characteristics, symbiogenesis, lateral gene transfer, etc) and call that the Extended Modern Synthesis.
I understand the motives, but there are several reasons why I did not opt for that way of presenting the change.
First, it is a central and specific feature of Neo-darwinism to exclude the inheritance of acquired characteristics. The Modern Synthesis took Darwin’s Natural Selection idea and added to it:
(1) the Neo-darwinist (NOT Darwinist) view that Lamarckism was impossible,
(2) that inheritance was entirely through Mendelian genes, and
(3) that variation in DNA is random in the sense that it is not directed by any functional processes.
In brief, the Neo-darwinian mechanism is that variations arise by chance, natural selection then works to see which of those variations win the competition to reproduce and dominate the gene pool of later generations. These assumptions are so fundamental to the Neo-darwinist view that it would be a strange hybrid to add to it precisely those mechanisms that it sought to exclude. It is more honest to say ‘we got it wrong’ by excluding them.
The analogy would be with the way in which Newtonian mechanics was replaced by relativity theory. It would have been absurd to call relativity theory Neo-newtonism! When experimental observations show precisely those processes that the theory did not predict, then we should say so. Somehow that needs to be recognised. Just as the Neo-darwinists saw themselves as developing a new theory by integrating Mendelian genetics with natural selection, we now need in some way to recognise that nature is even more wondrous than they thought, and involves processes we thought were impossible.
Second, it is important for historical reasons to recognise the injustices done to Lamarck, Waddington, McClintock, Margulis …….and many others. The dogmatic way in which Neo-darwinism was promulgated damaged reputations, it damaged careers, and it damaged whole disciplines which, like physiology, were excluded from contributing to the concepts of evolutionary biology. In my view, it even affected the reputation of Charles Darwin. Darwin was far from being a Neo-darwinist. He included the inheritance of acquired characteristics in his Origin of Species, even formulated his own mechanism (his theory of gemmules), and he acknowledged Lamarck in glowing terms: “this justly celebrated naturalist….who upholds the doctrine that all species, including man, are descended from other species.” (Preface to the 4th edition of The Origin of Species, 1866).
Third, there is the problem of nomenclature. Neo-darwinists, by using that term, captured the glow of Darwin’s name, but they do not always clearly distinguish whether they are talking about Darwinism or Neo-darwinism. This is the reason why criticisms of Neo-darwinism are often interpreted as criticism of Darwin. As Waddington knew well, it is perfectly possible to be a Darwinist without being a Neo-Darwinist. In the article I write:
“I start with some definitions. I will use the term ‘Modern Synthesis’ rather than ‘Neo-Darwinism’. Darwin was far from being a Neo-Darwinist (Dover, 2000; Midgley, 2010), so I think it would be better to drop his name for that idea. As Mayr (1964) points out, there are as many as 12 references to the inheritance of acquired characteristics in The Origin of Species (Darwin, 1859) and in the first edition he explicitly states ‘I am convinced that natural selection has been the main, but not the exclusive means of modification’, a statement he reiterated with increased force in the 1872, 6th edition.”
These considerations lead to the following conclusions:
- It would be better to drop the name ‘Neo-darwinism’ in describing what replaces it.
- The Modern Synthesis incorporates Neo-darwinism, which is why I incline towards dropping this name also. But, if we are to refer to an extended synthesis, Extended Modern Synthesis would be better than Extended Neo-darwinism since combining Neo-darwinism with Lamarckism would be a stark contradiction.
- I prefer the term ‘Integrative Synthesis’ since it highlights the fact that it would be an integration of many mechanisms, each playing roles whose importance can vary at different stages of the evolutionary process, and each of which can interact with the others. This would be a genuinely systems biological view of evolution, emphasising those interactions.
What replaces the Modern Synthesis will necessarily be a hybrid incorporating different mechanisms. I suspect that the only common feature will be that evolution happened.
These points can also be explained using a development of a valuable diagram from Pigliucci and Müller. Click on the movie below
The movie shows the transformation from Piggliucci and Müller’s figure to the one below:
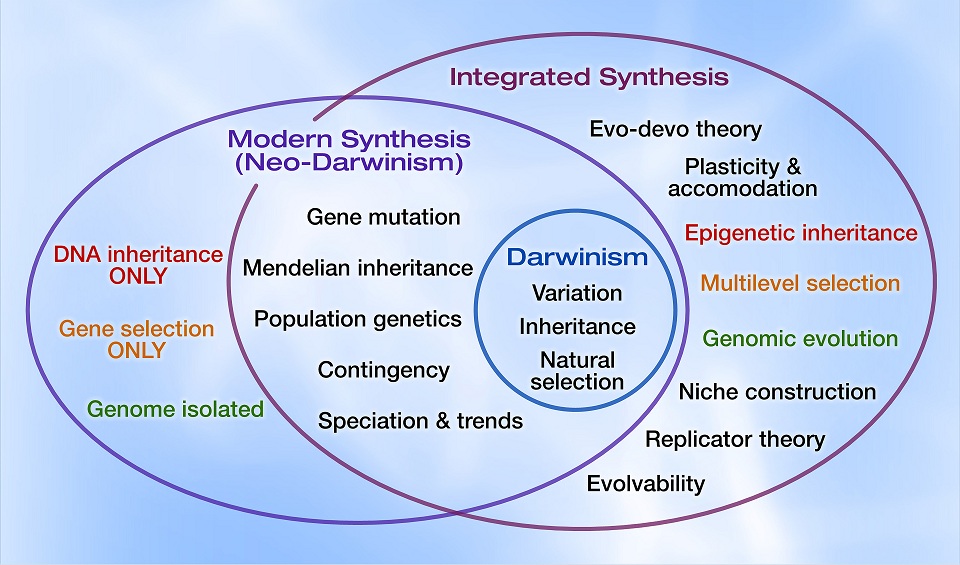
Diagram developed from Pigliucci, M., and Müller, G. B. (2010) Evolution – The extended synthesis, MIT Press, Cambridge, Mass.
The central circle represents the main features of Darwinism. The left hand ellipse represents most of the additional features of neo-darwinism (modern synthesis). The right hand ellipse represents the proposed extension.
My view is that the extension can best be represented as an extension of Darwinism. Nearly everything added is compatible with the original Darwinist assumptions. The main exception is that genomic evolution includes lateral transfer which modifies the Darwinian ‘tree of life’ to become a ‘network of life’.
The difficulties with representing the extension as one of neo-darwinism is that there are several negative characteristics of neo-darwinism that are not specifically referred to but which make it difficult from a historical point of view to see neo-darwinisn as the base for the extension. As explained above, neo-darwinism explicitly excluded the inheritance of acquired characteristics, denied the role of processes of genome change other than by chance, excluded symbiogenesis, and embraced the central dogma. All these need to be abandoned. The coloured items in my version of the diagram indicate some of the features that are in direct contradiction to neo-darwinism.
Since writing this page in 2013 I have been able to check with one of the authors of the Extended Evolutionary Synthesis who confirms that they also see it as a replacement not as a simple extension of neo-Darwinism.
Common (hilarious!) misunderstandings
The articles, lectures, and these webpages of answers have attracted considerable critical interest from various internet bloggers, most particularly from those who seem to display their Neo-darwinian dogmatism like a badge of honour. An unfortunately frequent feature of these blogs is that they mix the dogmatism with insulting, even libellous, language. See also dogmatism.
The best advice I can give to them and their readers is ‘look before you leap’. Firing off standard ready-made criticisms before carefully reading the articles or listening through the videos to the point at which those common criticisms are clearly dealt with simply perpetuates misunderstandings. In this section I deal with examples written by critics who are clearly ‘firing off from the hip’ since the points they make are already extensively dealt with in the lectures and articles. They only had to read or watch a bit further.
Darwin was wrong?
“Here we go again: someone arguing that DARWIN WAS RONG” (sic).
This comment could not be more wide of the mark since I argue that Darwin was (largely) right!
This kind of criticism illustrates a common tactic by some Neo-Darwinists, which is to ‘recruit’ Darwin to their cause. Darwin was not a Neo-Darwinist and he even admired Lamarck. It is a simple historical mistake to conflate Darwinism with Neo-Darwinism.
Random variation. One critic complains
“What we mean by “random” is that mutations occur regardless of whether they would be good for the organism.”
Precisely so, and if the writer had read on just a little bit further (at the end of the same paragraph in which ‘random’ is first mentioned!) he would find my precisely equivalent statement: “I will use the definition that the changes are assumed to be random with respect to physiological function and could not therefore be influenced by such function or by functional changes in response to the environment. This is the assumption that excludes the phenotype from in any way influencing or guiding genetic change.”
The same ‘rapid fire’ mistake through not reading carefully is made by another blogger who wrote
“His most moronic claim by far is the one on mutations not being random.”
The extraordinary feature of this kind of criticism is that the potentially functional nature of some of the variations is the central theme of the articles and lectures. It is hard to miss that theme if one reads the article even cursorily, and it features towards the beginning of the IUPS2013 lecture. At 7:00 minutes the transcript reads “It is important here to ask what we mean by random, because it is not just a question whether the changes are truly random … but rather whether the changes are functionally relevant. That is the key.” At 7:30 minutes the transcript even uses the definition in the published article exactly as quoted above. It forms one of the slides of the lecture. In the 2012 lecture in Suzhou, China, it is made clear at 6:48 minutes and again towards the end of the lecture that the key lies in functionally significant changes. At 34:19 the transcript reads “If functional changes in the adult can be inherited, and therefore a target for natural selection, then physiology – which is the analysis of function – IS highly relevant to evolution.” It is hard to see how these points could have been made any clearer in the article and lectures. Failure to notice them implies a failure to read or watch carefully enough.
Notice again the gratuitous insults. There are even more grossly insulting and libellous remarks on the blog.
Inheritance of acquired characteristics.
“I know of not a single adaptation in organisms that is based on such environmentally-induced and non-genetic change.”
It is hard to take this kind of comment seriously. It reveals someone who is not keeping up with the literature. See Trans-generational inheritance for examples.
“For an adaptation to become fixed in a population or species, it must be inherited with near-perfect fidelity. And that is not the case for all environmentally-induced modifications of DNA. They eventually go away.”
I give examples in the lectures and article where this is clearly not true. RNA transmitted changes independent of DNA have been followed in planarians for 100 generations:
Rechavi O, Minevish G & Hobert O (2011). Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans. Cell 147, 1248–1256.
The recent Apobec1 deficiency example from Nadeau’s laboratory is also very clear in showing “heritable epigenetic changes [that] persisted for multiple generations and were fully reversed after consecutive crosses through the alternative germ-lineage.”
Nelson VR, Heaney JD, Tesar PJ, Davidson NO & Nadeau JH (2012). Transgenerational epigenetic effects ofApobec1 deficiency on testicular germ cell tumor susceptibility and embryonic viability. Proc Natl Acad Sci U S A 109, E2766–E2773
Notice also the very high quality of the journals (Cell and PNAS) in which these ground-breaking studies were published.
From a much earlier period (mid 20th century), the environmentally-induced changes investigated by Waddington became locked into the DNA after about 14 generations and therefore became essentially permanent. He called this process genetic assimilation and it was one of Waddington’s great, but largely ignored, contributions.
See Trans-generational inheritance for further examples and references. To quote my article: Some of “these effects persist for many generations and are as strong as conventional genetic inheritance.”
Cells are transitory.
“Cells are transitory, and DNA is not.”
This is a common mantra, copied from The Selfish Gene. It is linguistically incoherent and factually incorrect. See Immortal Genes?
Jumping genes.
“These kinds of changes are rare except in bacteria.”
Barbara McClintock received her Nobel Prize for jumping genes in 1983 for her work on plants. The examples I give in the lecture show large-scale genome reorganisation across a whole range of vertebrates. Cross-species transfers are indeed rare except in bacteria. But, after all, speciation itself is rare, and there are plenty of documented examples of horizontal gene transfer from bacteria to various eukaryotes. Recent examples include:
Redrejo-Rodríguez, M, Muñoz-Espín, D, Holguera, I, Mencía, M, Salas, M, (2012). Functional eukaryotic nuclear localization signals are widespread in terminal proteins of bacteriophages. Proc. Natl. Acad. Sci. U.S.A. 109: 18482–7. “These findings show a common feature of TPs from diverse bacteriophages targeting the eukaryotic nucleus and suggest a possible common function by facilitating the horizontal transfer of genes between prokaryotes and eukaryotes.”
Acuna, R. et al (2012) Adaptive horizontal transfer of a bacterial gene to an invasive insect pest of coffee. Proc. Natl. Acad. Sci. U.S.A. 109: 4197–4202. “We identified a gene (HhMAN1) from the coffee berry borer beetle, Hypothenemus hampei, a devastating pest of coffee, which shows clear evidence of HGT from bacteria.”
Where the phenomenon is common (in prokaryotes) speciation is so rapid that microbiologists no longer find the species concept helpful.
The more (and only) serious point made by this particular critic is whether the Modern Synthesis has been modified to take account of these ‘big mutations’:
“The Modern Synthesis has expanded a bit to take account of these new genetic findings, which only recently became possible. But their discovery hardly invalidates the Synthesis.”
I think that is a matter of judgment. Some of the new findings are incompatible with the Modern Synthesis.
Origin of species. A common criticism is that speciation has been observed. Precisely so, and I don’t question that. The relevant question now is what the mechanisms were. It is common in Neo-darwinist discourse for people to conflate the fact that speciation clearly has and does occur with the specific Neo-darwinist interpretation of the mechanism by which it happens. This is part of a pattern in which any evidence for the fact of evolution is counted as evidence for Neo-darwinism. To say the least, this is sloppy thinking.
And, finally……..
Good advice to Denis Noble.
“He might try discussing his ideas with other evolutionists and listening to their responses.”
I couldn’t agree more! I have greatly enjoyed doing so for around 40 years. The list of leading Evolutionary Biologists with whom I have interacted must now be around fifty, and they involve some well-known Neo-darwinists, including Dawkins, Maynard Smith and Wolpert.
Conclusions
The critical reactions so far are so extraordinarily wide of the mark that they are simply hilarious – almost a parody of academic criticism. Yet they weren’t intended to amuse, they were meant in all seriousness.
They attack positions that it is obvious I do not hold. That could have been clear from reading the articles or viewing the lectures. If the critics did that, they must have done so with remarkably closed minds or they failed to follow the logic through to their clearly stated conclusions. To miss the main statement when it is in the same paragraph as the sentence being attacked is breath-taking. It is an elementary principle of scholarship to read an article fully before writing a criticism.
The need to use abusive libellous language also speaks volumes about the confidence these people must have in their own scholarship.
So, are these critics just ‘saloon bar’ opinionators, or teenagers having fun on the internet? No, they are full university professors at major universities. And they are serious. That is what is both interesting and alarming, and why I have taken the time to answer them seriously. But how could such a major area of science have generated such philosophically naïve dogmatism?
Common-Variant Common-Disease Hypothesis
Contributed by Michael Joyner
One widely assumed outcome of the Human Genome Project (HGP) was that a limited number of gene variants based on so-called single nucleotide polymorphisms (SNPs) would explain much of the risk of common non-communicable diseases like ischemic heart diseases, type II diabetes, hypertension and other conditions including some forms of cancer. This is known as Common-Variant Common-Disease Hypothesis and was underpinned by the well-known observation that many diseases run in families and are heritable in a statistical sense. However, these conditions do not have classic Mendelian patterns of heritability, as for example, certain forms of hemophilia, sickle cell anemia, or cystic fibrosis do. In this context, the results of numerous Genome Wide Association Studies (GWAS) conducted in large populations of humans have found:
- A large number of gene variants (sometimes hundreds) are associated with many common diseases.
- The effect size of such variants is typically small.
- The reproducibility GWAS studies from one study or population to the next can be problematic.
- The distribution of variants in populations with and without a given disease is frequently similar.
- The addition of gene scores to disease risk prediction algorithms derived from epidemiology studies has generally not improved the algorithms.
These findings highlight the ongoing problem of “missing heritability” which has been a concern since studies from the Victorian era on the relationship between the height of parents and their children. For those who favor a DNA centric view of human phenotypic variability the assumption is that more detailed studies of the genome will shed light on DNA based mechanisms that might account for this missing heritability. Those who favor the view that human phenotypic variability is due to complex interactions between the environment, behavior, physiological regulation and adaptation, and the genome see the continuing challenge of missing heritability as highlighting the limitations of the DNA centric view of human phenotypic variation.
For more information see:
Baker M (2010). Genomics: The search for association. Nature 467: 1135-1138. http://www.nature.com/nature/journal/v467/n7319/full/4671135a.html
Collins FS (2001). Contemplating the end of the beginning. Genome Res 11: 641-643. http://genome.cshlp.org/content/11/5/641.full
Cooper GM, Shendure J (2011). Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat Rev Genet. 12:628-40. http://www.nature.com/nrg/journal/v12/n9/full/nrg3046.html
Ganesh SK, et al Genetics and genomics for the prevention and treatment of cardiovascular disease: update: a scientific statement from the American Heart Association. Circulation 128: 2813-2851. http://circ.ahajournals.org/content/128/25/2813
Joyner MJ, Prendergast FG (2014). Chasing Mendel: five questions for personalized medicine. J. Physiol in Press.
Shields R (2011). Common disease: are causative alleles common or rare? PLoS Biol 9: e1001009. http://www.plosbiology.org/article/info%3Adoi%2F10.1371%2Fjournal.pbio.1001009
Talmud PJ et al (2010). Utility of genetic and non-genetic risk factors in prediction of type 2 diabetes: Whitehall II prospective cohort study. Br Med J 340: b4838. http://www.bmj.com/content/340/bmj.b4838
Does the 2013 article criticise Darwinism?
No. Not really. In fact the main thrust of the article is a return to a less dogmatic view which is more in keeping with Darwin’s original ideas.

“In some respects, my article returns to a more nuanced, less dogmatic view of evolutionary theory (see also Müller, 2007; Mesoudi et al. 2013), which is much more in keeping with the spirit of Darwin’s own ideas than is the Neo-Darwinist view.”
Müller GB (2007). Evo–devo: extending the evolutionary synthesis. Nat Rev Genet 8, 943–949. http://www.ncbi.nlm.nih.gov/pubmed/17984972
Mesoudi A, Blanchet S, Charmentier A, Danchin E, Fogarty L, Jablonka E, Laland KN, Morgan TJH, Mueller GB, Odling-Smee FJ & Pojol B. (2013). Is non-genetic inheritance just a proximate mechanism? A corroboration of the extended evolutionary synthesis. Biological Theory 7, 189–195. http://link.springer.com/article/10.1007/s13752-013-0091-5
“One of the major developments of Darwin’s concept of a ‘tree of life’ is that the analogy should be more that of a ‘network of life’ (Doolittle, 1999; Woese & Goldenfeld, 2009).”
DoolittleWF (1999). Phylogenetic classification and the universal tree. Science 284, 2124–2128. http://www.ncbi.nlm.nih.gov/pubmed/10381871
Woese CR & Goldenfeld N (2009). How the micobial world saved evolution from the Scylla of molecular biology and the Charybdis of the modern synthesis. Microbiol Mol Biol Rev 73, 14–21. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2650883/
The network represents the evidence for extensive exchange of DNA that must have occurred in the early stages of evolution, but which also continued through later stages.
The main departure from Darwin’s ideas is that the ‘tree of life’ is a network:
What explains the dogmatism and persistence of neo-darwinism?

When the history of neo-Darwinism gets written it will need to answer this question. I am not a historian. The answers I give here are tentative. They could be leads for serious historians of science to follow up. I also highlight two brilliant scientists, Waddington and McClintock, whose work should not have been neglected.
The progress of science is not Popperian
Despite the great influence of Popper, single contrary observations rarely destroy a strongly established theory. The tendency is to fix theories, extend them, even to redefine their entities, in ways that allow the contrary observations to be absorbed. This is what happened to Waddington’s work. If they can’t be absorbed in this way, they are sidelined as anomalies. This nearly happened to McClintock until she was awarded the Nobel Prize for discovering mobile genetic elements (jumping genes). It is more often the accumulation of many forms and examples of contrary evidence that persuades people to change. I think we are at that point now in the case of neo-Darwinism. There are exceptions to all the central assumptions, and in the case of the inheritance of acquired characteristics the exceptions are becoming a flood of evidence. Moreover, the central element of the theory, the gene, is no longer easy to define. All those functional RNAs also act as ‘genes’. The marking of the genome and chromatin also play a role in inheritance. That is the reason why a growing number of scientists admit that a major rethink is already happening.
Conrad Waddington in 1946 (left, from The Royal Society picture library) and his diagram of the epigenetic landscape (right, from The Strategy of the Genes, 1957). Genes (solid pegs at the bottom) are viewed as parts of complex networks so that many gene products interact to produce the phenotypic landscape (top) through which development occurs. Waddington’s insight was that new forms could arise through new combinations to produce new landscapes in response to environmental pressure, and that these could then be assimilated into the genome. Waddington was a systems biologist in the full sense of the word. If we had followed his lead many of the more naïve 20th century popularizations of genetics and evolutionary biology could have been avoided.

Left: Barbara McClintock in her laboratory in 1947 (Smithsonian Institute Archives).
“In the future attention undoubtedly will be centered on the genome, and with greater appreciation of its significance as a highly sensitive organ of the cell, monitoring genomic activities and correcting common errors, sensing the unusual and unexpected events, and responding to them, often by restructuring the genome. We know about the components of genomes that could be made available for such restructuring. We know nothing, however, about how the cell senses danger and instigates responses to it that often are truly remarkable.” (McClintock, Barbara. 1984 The significance of responses of the genome to challenge. Science 226, 792-801.)
McClintock clearly understood the correct direction of causality in relation to the genome. Characterising the genome as “a highly sensitive organ of the cell” was her great insight. It is as foolish to regard the genome as the active cause of the organism as it would be to say that the pipes of an organ play the music!
Right: giving Nobel Prize lecture in 1983 (NIH)
Cold War
Theories of evolution became an issue during the Cold War. Those, like Waddington, who found evidence for the inheritance of acquired characteristics were thought in some way to be associated with Lysenkoism, a Soviet era school that denied Mendelian inheritance. Waddington was not even allowed to visit the USA. His work was virtually ignored in the USA, and was eventually sidelined in the UK. He saw himself as a Darwinist, but not as a neo-Darwinist. I think it is shameful that such a brilliant scientist was side-lined, just as I think it is shameful that Lamarck was virulently denigrated. Anyone who thinks that science is neutral, and is not influenced by politics, should answer the question: how did these injustices happen?
See also http://musicoflife.website/pdfs/LetterfromLamarck.pdf

Religious fundamentalism
It is said that nearly 50% of the population of the USA do not accept the theory of evolution. Some are called creationists since they believe in various forms of creation, either literally as described in Genesis, or in a variety of more modern ideas of creationism. Some also espouse the ideas of Intelligent Design (ID). Both the creationists and the supporters of ID tend to take every example of a break with neo-Darwinism as a vindication of their views. Some have done the same with my article, despite the fact that I make it clear that I am arguing for a return to a “more nuanced, less dogmatic view of evolutionary theory (see also Muller, 2007; Mesoudi ¨ et al.2013), which is much more in keeping with the spirit of Darwin’s own ideas than is the Neo-Darwinist view.”
Muller GB (2007). Evo–devo: extending the evolutionary synthesis. Nat Rev Genet 8, 943–949 http://www.ncbi.nlm.nih.gov/pubmed/17984972
Mesoudi A, Blanchet S, Charmentier A, Danchin E, Fogarty L, Jablonka E, Laland KN, Morgan TJH, Mueller GB, Odling-Smee FJ & Pojol B. (2013). Is non-genetic inheritance just a proximate mechanism? A corroboration of the extended evolutionary synthesis. Biological Theory 7, 189–195. http://link.springer.com/article/10.1007/s13752-013-0091-5
One way to view the dogmatic nature of neo-darwinism as it is often presented in public is to see it as a reaction to the dogmatism of the creationists. The ‘uncertain’ (in the sense of lacking reason) faith in creationism is replaced by the ‘certainties’ of science. But there is a conflation here of very different degrees of certainty in science. There can’t be much doubt about the fact that life on earth has evolved. There is much less certainty about the mechanisms. Unlike Darwinism (Darwin knew nothing of mechanisms, genes were not known), neo-darwinism proposes the exclusion of many mechanisms that have in fact now been found to occur in nature. Adopting the ‘certainty’ of evolution to clothe the ‘uncertainty’ of particular theories about mechanisms has been the cause of many problems in public debate on evolution. It is perfectly possible to defend the virtual certainty that life has evolved while debating in the usual argumentative scientific way the uncertainties surrounding the question of mechanisms. The truth is that amongst the many mechanisms now known we know very little about which were prevalent in evolution. The answer is likely to be that different mechanisms were dominant at different stages. Evolution itself evolves.
See also the item on The Language of Neo-darwinism where the discourse of neo-darwinism is analysed.
What does DNA do?
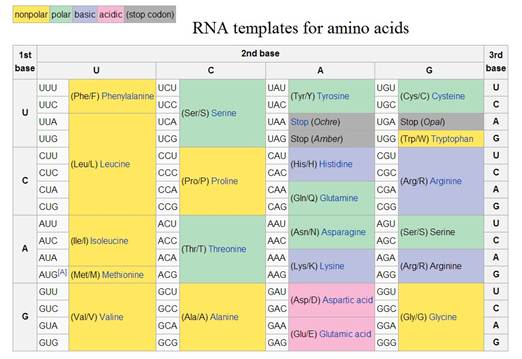
Table of the nucleic acid template patterns for the formation of amino acids in protein sequences (from wikipedia). These triplet patterns are formed from the four bases U (uracil in RNAs), T (thymine in DNA), C (cytosine), A (adenine), and G (guanine), and are often described as the genetic ‘code’, but it is important to understand that this usage of the word ‘code’ is metaphorical and can be confusing. A real code is an intentional encryption used by humans to communicate. The genetic ‘code’ is not intentional in that sense. The more neutral word ‘template’ is better. It also expresses the fact that templates are used only when required (activated), they are not themselves active causes.
DNA sequences are templates for the formation of RNAs, some of which are in turn used as templates for the formation of proteins. Only RNAs and proteins are specified by the DNA sequences. RNAs and proteins then cooperate with many metabolites and other components of the organism to generate the numerous interlocking biochemical pathways. These pathways in turn work within the three dimensional structure of the cells, tissues and organs of the body. But this three-dimensional structure is not to be found in the one-dimensional DNA sequences. How then do cells know how to form such an intricate biological mechanism of precise functionality? The answer is self-templating. The three-dimensional structures copy themselves, first by growing in size and then, when the cell becomes large enough to divide, the structures are divided between the two cells. Each daughter cell therefore inherits its complete complement of structure as well as inheriting the genome.
For further aspects of the answer to this question see Immortal genes?
Why should physiologists be concerned about these questions?
A major problem with the modern synthesis from the viewpoint of physiology is that it excludes phenotypic function from having any role whatsoever in influencing the direction and frequency of genomic change. This is what neo-darwinists really mean when they refer to gene changes as ‘random’. It doesn’t really matter to them whether the changes are ‘truly’ random. What matters is whether function can influence those changes. Through that crack, if it exists, will flow all that they wish to exclude, including strong forms of the inheritance of acquired characteristics. If the modern synthesis is correct, then physiology really is dealing with the disposable carrier of genes. If however, on this central point, it is incorrect then, as I say in the article “It is hard to think of a more fundamental change for physiology and for the conceptual foundations of biology in general”. That, incidentally, is the justification for the title of the article. I did not choose the title light-heartedly.
PLEASE NOTE that the answer to this very important question is necessarily very technical. But I think it is necessary to give the full details. The angel in this case lies in the detail. The devil will lie with those who are unprepared to study the detail but who still wish to claim that the ‘earth has not moved’.
The key to this question lies in four major discoveries:
First, some of the non-random changes in the genome are functionally significant.
“Changes in the speed of change are well known already from the way in which genome change occurs in immunological processes. The germ line has only a finite amount of DNA. In order to react to many different antigens, lymphocytes ‘evolve’ quickly to generate extensive antigen-binding variability. There can be as many as 1012 different antibody specificities in the mammalian immune system, and the detailed mechanisms for achieving this have been known for many years. The mechanism is directed, because the binding of the antigen to the antibody itself activates the proliferation process. Antigen activation of B-cell proliferation acts as a selective force.”
This example was given first because targeted genome change in the immune system is well-documented and has been known for a long time. That it can happen in B-cells as they ‘evolve’ to generate the variability shows that the mechanism of such targeted genome change is not new. We should not therefore be surprised to find that it is used elsewhere in the organism. Now let’s move to the part of the article that deals with evidence beyond the immune system.
“Similar targeted genomic changes occur outside the context of the immune system. The reader is referred to table II.7 (Shapiro, 2011, pp. 70–74;
http://shapiro.bsd.uchicago.edu/TableII.7.shtml) for many examples of the stimuli that have been shown to activate this kind of ‘natural’ genetic engineering, while table II.11 from the same book documents the regions of the genomes targeted. (pp. 84–86; http://shapiro.bsd.uchicago.edu/TableII.11.shtml). Thirty-two examples are given. One example will suffice to illustrate this. P element homing in fruit flies involves DNA transposons that insert into the genome in a functionally significant way, according to the added DNA. There is up to 50% greater insertion into regions of the genome that are related functionally to DNA segments included within the P element. Thus, ‘Insertion of a binding sequence for the transcriptional regulator Engrailed targets a large fraction of insertions to chromosomal regions where Engrailed is known to function.’ (Shapiro, 2011, p. 83)”
The reference and abstract for this example is
Cheng, Y., Kwon, D. Y., Arai, A. L., Mucci, D., and Kassis, J. A. (2012) P-Element Homing Is Facilitated by engrailed Polycomb-Group Response Elements in Drosophila melanogaster PLoS ONE 7, 1. http://www.ncbi.nlm.nih.gov/pubmed/22276200
ABSTRACT: P-element vectors are commonly used to make transgenic Drosophila and generally insert in the genome in a non-selective manner. However, when specific fragments of regulatory DNA from a few Drosophila genes are incorporated into Ptransposons, they cause the vectors to be inserted near the gene from which the DNA fragment was derived. This is called P-element homing. We mapped the minimal DNA fragment that could mediate homing to the engrailed/invected region of the genome. A 1.6 kb fragment of engrailed regulatory DNA that contains two Polycomb-group response elements (PREs) was sufficient for homing. We made flies that contain a 1.5kb deletion of engrailed DNA (enΔ1.5) in situ, including the PREs and the majority of the fragment that mediates homing. Remarkably, homing still occurs onto the enΔ1.5 chromosome. In addition to homing to en, P[en] inserts near Polycomb group target genes at an increased frequency compared to P[EPgy2], a vector used to generate 18,214 insertions for the Drosophila gene disruption project. We suggest that homing is mediated by interactions between multiple proteins bound to the homing fragment and proteins bound to multiple areas of the engrailed/invected chromatin domain. Chromatin structure may also play a role in homing.
The end of this abstract echoes another point I make in the article:
“Structural organization also represents information that is transmitted down the generations. DNA is not merely a one-dimensional sequence. It is a highly complex physiological system that is regulated by the cells, tissues and organs of the body.”
Finally, another well-known functionally-driven form of genome change is the response to starvation in bacteria.
“An important point to note is the functionally significant way in which this communication can occur. In bacteria, starvation can increase the targeted transposon mediated reorganizations by five orders of magnitude, i.e. by a factor of over 100,000 (Shapiro, 2011, p. 74).”
Second, some forms of non-DNA (epigenetic) inheritance have been shown to be as robust as DNA inheritance and to be transmitted for many generations.
The details on this point are given in the answer to the question how widespread non-DNA inheritance is.
Third, inherited changes that occur by whatever mechanism can become locked into the genome by genetic assimilation. This was the major conclusion of Waddington’s experiments in fruit flies – see
Bard JBL (2008). Waddington’s legacy to developmental and theoretical biology. Biological Theory 3, 188–197. http://www.deepdyve.com/lp/mit-press/waddington-s-legacy-to-developmental-and-theoretical-biology-o0mS0JjRau
Waddington showed a form of inheritance of an acquired characteristic that was initially ‘soft’ in the sense that it required repetition of the environmental stimulus in each generation to maintain it. But after about 14 generations it became ‘hard’, i.e. assimilated into the genome. I think that what was happening was that the separate alleles necessary for the characteristic were already present in the population but not in the right combination to be expressed without the stimulus. The environmental stimulus was eventually not needed because by that generation the correct combination of alleles was now present in individuals who could pass this pattern of alleles on to the next generation in the standard genetic way. I summarise this point in the article by writing “After all, the pattern of the genome is as much inherited as its individual components, and those patterns can be determined by the environment.”
Fourth, some trans-generational effects can occur reliably by bypassing the genome.
“Epigenetic effects can even be transmitted independently of the germ line. Weaver and co-workers showed this phenomenon in rat colonies, where stroking and licking behaviour by adults towards their young results in epigenetic marking of the relevant genes in the hippocampus that predispose the young to showing the same behaviour when they become adults (Weaver et al. 2004; Weaver, 2009).”
Weaver ICG, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M & Meaney MJ (2004). Epigenetic programming by maternal behavior. Nat Neurosci 7, 847–854 http://www.ncbi.nlm.nih.gov/pubmed/15220929
Weaver ICG (2009). Life at the interface between a dynamic environment and a fixed genome. In Mammalian Brain Development. ed. Janigro D, pp. 17–40. Humana Press, Springer, New York, NY, USA.
Neo-darwinists tend to dismiss this kind of example as a form of cultural inheritance. So it is. But it works by marking the genome of the next generation. It is therefore just as relevant as and just as robust as epigenetic inheritance in general. Rats and other rodents do it all the time.
Jonny-cum-lately?
This English expression refers to someone who enters an activity, such as a field of study, or a competition, very late and then tries to run off with the prizes. The implication is that he/she has no right to do so since they didn’t do the hard work earlier.
I use this expression as the title of this item because, amongst the more unbelievable critical comments on the internet, is one that tries to dismiss me as someone who has no track record in the field and who has only entered it very late in his career. This could not be further from the truth. I have worked on these issues for over 40 years. I organised what was probably the first debate on The Selfish Gene way back in 1976, the year of its publication. The participants were Richard Dawkins, Anthony Kenny, Charles Taylor and me. I have frequently taken part in Novartis Foundation discussion meetings on these questions, including defenders of the Modern Synthesis such as Lewis Wolpert and John Maynard Smith. At the 1993 IUPS Congress in Glasgow, where I was chairman of the organising committee, I edited The Logic of Life: the challenge of integrative physiology (OUP, 1993) in which I also co-authored the lead chapter. As the congress opened I published a full page article in the national Sunday newspaper, the Observer, entitled Unravelling the meaning of life. A key quotation from that article is
“Could we have mistaken identifying the code for the mechanics of living cells for an understanding of the logic of life itself?”
An article by Tim Radford, the chief Science correspondent of The Guardian, just 2 days previously refers to me as having “turned the idea of the selfish gene on its head.” The central idea of The Music of Life and my quarrel with Neo-Darwinism is more than 20 years old.
As I show in a more recent book, the gestation period of The Music of Life (OUP 2006) was many years. It would have been impossible to write such a book without the experience I have just outlined.
The same is true of the article I wrote in 2011 deconstructing The Selfish Gene and Neo-Darwinism. Anyone who reads that article can see the evidence of years of work on the analysis.
People don’t come ‘out of the blue’ to write an article with over 80 detailed references without such years of experience.
What is a template?
In the item The Language of Neo-darwinism I refer to DNA sequences as templates rather than as code. What is a template and why do I think that is a better metaphor?
I am the fortunate owner of three classical guitars made by the top English luthiers, David Rubio and Paul Fischer. David Rubio made guitars for Julian Bream and the label of one of my Rubios is signed by Bream. I inherited this guitar by buying it from my guitar teacher.
Guitar made by Paul Fischer with uniform fine-grained cedar for the soundboard

Needless to say, these guitars are a joy to play. I am not really a good enough performer for them but I love being able to play them. Like inheriting a good genome from your parents, it helps with life, it gives you a head start!
Why are these guitars so good? All top luthiers select the wood very carefully and season it correctly. The soundboard must have an almost perfectly uniform grain. But this is not all. The soundboard resonance properties are also determined by struts that are placed on the underside of the wood, inside the guitar and hidden from view. The pattern of these struts is unique to each luthier. In their striving to combine sweet tone with increased volume, they experiment extensively with exactly how the struts should be placed.
Recently, I acquired the template that Paul Fischer used to ensure that his struts follow his designed pattern. He has now retired from guitar making and no longer needs it. The template is for a fan strut pattern, which is characteristic of his guitars


The precise placing of these fan struts (braces) determines the balance between the resonances of the wood at different frequencies. In a sense therefore the template used is a cause of the way in which the instrument plays. So also is the way in which the guitarist strikes the strings. Even the shape, length and smoothness of the fingernails are important. But clearly we are dealing here with different kinds of cause. The template is a passive cause. Its existence and use were necessary but in no sense can the template be said to play the music. The active cause of the music that the instrument produces is the act of playing by the guitarist. This distinction between active and passive causes is important.
This is the distinction that is ignored when people say that their genes ‘made them … body and mind’. The genome is our template. Our protein networks within the cells and organs of the body are our guitar. But we play the music of life.
The genome sequences work just like Fischer’s template. They determine how my body’s proteins are structured. But they do not themselves carry out the physiological functions served by proteins, metabolites and the three-dimensional cellular and organ structures within which they work.
It’s as simple as that.

With thanks to Paul Fischer
Luthier extraordinaire
Barker Hypothesis, maternal fetal programming and human health
Contributed by Michael Joyner
Epidemiologic and anthropological studies in humans have shown that maternal (and thus fetal) under or over nutrition can have implications for the lifelong health of the offspring especially as it relates to metabolic regulation and blood pressure. For example fetal under or over nutrition can increase the risk of obesity, hypertension and diabetes in the offspring especially if they are subsequently exposed to an environment with plentiful food. Importantly, these effects can be multigenerational even in the absence of in utero nutritional stress. Additionally, these population based observations have been confirmed in animal models. These multigenerational effects are not the result of specific changes in the sequences of DNA that code for proteins important in the regulation of metabolism and blood pressure. They highlight the limitations of the DNA centric view of human phenotypic variation and are a key example of soft inheritance. They emphasize the idea that complex multidirectional interactions between the environment, behavior, physiological regulation and adaptation, and the genome explain human phenotypic variation.
For more information see:
http://www.thebarkertheory.org/
Calkins K, Devaskar SU (2011). Fetal origins of adult disease. Curr Probl Pediatr Adolesc Health Care. 41:158-76. http://www.ncbi.nlm.nih.gov/pubmed/16368278
Yes. The 2013 article criticises neo-Darwinism (the modern synthesis) as being too restrictive and too dogmatic. That view becomes a special case that holds under certain circumstances.
“As with other breaks from the Modern Synthesis, that synthesis emerges as only part of the evolutionary story.”
The modern synthesis is also criticised for the unwarranted and virulent denigration of Lamarck.
“In 1998, the great contributor to the development of the Modern Synthesis, John Maynard Smith, made a very significant and even prophetic admission when he wrote ‘it [Lamarckism] is not so obviously false as is sometimes made out’ (Maynard Smith, 1998), a statement that is all the more important from being made by someone working within the Modern Synthesis framework. The time was long overdue for such an acknowledgement. Nearly 50 years before, Waddington had written ‘Lamarck is the only major figure in the history of biology whose name has become to all extents and purposes, a term of abuse. Most scientists’ contributions are fated to be outgrown, but very few authors have written works which, two centuries later, are still rejected with an indignation so intense that the skeptic may suspect something akin to an uneasy conscience. In point of fact, Lamarck has, I think, been somewhat unfairly judged.’ (Waddington, 1954).”
Maynard Smith J (1998). Evolutionary Genetics. Oxford University Press, New York, NY, USA.
Waddington CH (1954). Evolution and epistemology. Nature 173, 880–881.
The inheritance of acquired characteristics, usually called Lamarckism, has now been demonstrated, it can persist for many generations, and some of the molecular mechanisms of such inheritance have been found. It is time that this was openly acknowledged.
The reason that the new developments are ‘rocking the foundations’ is that it was clearly an aim of the modern synthesis to exclude the inheritance of acquired characteristics. It is precisely that aspect of the modern synthesis that has now been shown to be incorrect.
The question whether neo-Darwinism has been proven wrong therefore depends on what exactly is being questioned. The dogmatic claims (a) that the inheritance of acquired characteristics is impossible, (b) that all evolutionary change is incremental accumulation of ‘random’ mutations, (c) that the tree of life does not include lateral transfer to form a network of life, have quite clearly been disproven by experimental work. I can’t understand why neo-darwinists cannot accept this. But we must also avoid the reverse dogma: the neo-darwinist view of evolutionary mechanisms has not been disproven. It has simply become one of several mechanisms of evolutionary change.
Does evolution evolve?
It must have done so.
The neo-darwinist process is a late comer since it is unlikely to have been the main mechanism before the development of multicellular organisms with separate specialised germline cells. It is a zoology-orientated view of evolution. Zoological organisms have existed for only about 20% of the duration of life on earth.
Similarly, inheritance via DNA cannot have occurred before the evolution of DNA. Sexual selection and mixing mechanisms cannot have evolved before the mechanisms for fusing, dividing and recombining genomes.
The evolution of the cell probably occurred before that of DNA, and certainly before DNA organised into genomes.
In fact, the list of stages is almost endless. The organisation of genomes, particularly genomes with chromatin and enclosed in a nucleus, must have occurred in many stages.
Not only does evolution evolve, so does the context in which it happens since it also changes the environment. New niches are created by evolution which can then be the context for new forms to evolve. It is an endless wondrously intricate web of contexts, structures and mechanisms. It can’t be reduced to a simple formula with one paradigmatic mechanism.
Laland, KN, Odling-Smee, J, Feldman, MW, Kendal, J. (2009) Conceptual Barriers to Progress Within Evolutionary Biology. Found Sci 14, 195-216 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3093243c
Odling-Smee, F. J., Erwin, D., Palkovacs, E. P., Feldman, M. W., and Laland, K. N. (2013) Niche Construction Theory: a practical guide for ecologists, The Quarterly Review of Biology 88, 4-28. http://www.ncbi.nlm.nih.gov/pubmed/23653966
“It’s highly plausible, it’s economical, it’s parsimonious, why on earth would you want to drag in symbiogenesis when it’s such an unparsimonious, uneconomical [theory]?”
Margulis replied:
“Because it’s there.”
That’s it in a nutshell. What is there, what exists, which is the starting point of all science.
See
http://www.voicesfromoxford.org/video/Homage-to-Darwin-part-1/74
http://www.voicesfromoxford.org/video/Homage-to-Darwin-part-2/63
http://www.voicesfromoxford.org/video/Homage-to-Darwin-part-3/75
for the debate
http://musicoflife.website/pdfs/HOMAGE_COMMENTARY_Music%20of%20Life.pdf for a commentary

An image from the series of three videos recording the 2009 debate on the Voices from Oxford website. Click on the image to access the videos.
See also Two Wrongs for more information on Lynn Margulis and the way in which her major contributions to the theory of evolution have been denigrated. Yet the transition to cells with mitochondria or chloroplasts was surely an absolutely fundamental step in evolution.
Immortal genes?
The DNA passed on from one generation to the next is a copy (though not always perfect). The cell that carries the DNA is also a copy (also not always perfect). In order for a cell to give rise to daughter cells, both the DNA and the cell have to be copied (replicated – ‘replicate’ means ‘to make a copy’). The only difference between copying a cell and copying DNA is that the cell copies itself by growing (copying its own detailed structure gradually, which is an example of self-templating) and then dividing so that each daughter cell has a full complement of the complex cell machinery and its organelles, whereas copying DNA for the purpose of inheritance occurs only when the cell is dividing.
Moreover, the complexity of the structure in each case is comparable. See Noble (2011) Differential and Integral views of genetics, particularly page 9: “It is therefore easy to represent the three dimensional image structure of a cell as containing as much information as the genome.”
My germ line cells are therefore just as much ‘immortal’ as their DNA. Moreover, nearly all of my cells and DNA die with me. Those that do survive, which are the germ cells and DNA that help to form the next generation, do not do so separately. DNA never works without a cell. It is simply an incorrect playing with words to single the DNA out as uniquely immortal.
I was also playing with words when I wrote that “DNA alone is inert, dead.” But at least that has a point. DNA alone does nothing. Cells can continue to function for some time without DNA. Some cells do that naturally, e.g. red blood cells which live for about 100 days without DNA. Others, such as isolated nerve axons or any other enucleated cell type, can do so in physiological experiments.
The point I am making is that functionality lies with the cell. DNA is an inert set of templates that the cell uses to make proteins and RNAs. Genes are therefore causes in a passive sense. They do nothing until activated. A set of proteins then initiates the process of transcribing the relevant templates. Active causation lies with proteins, membranes and the active functional networks they form. See Noble (2008) Phil Trans Roy Soc A, 366, 3001-3015
Distinguishing between the various senses of ‘cause’ is an elementary principle of philosophy. See chapter 1 of The Music of Life.
Jean-Baptiste Pierre Antoine de Monet, Chevalier de la Marck
Jean-Baptiste Pierre Antoine de Monet, Chevalier de la Marck This is the person usually called Lamarck. He was born in 1744, before the French Revolution. He became Professor at the famous Jardin des Plantes in Paris, where the museums contained a rich collection of geological objects gathered from all over the world. He was initially strongly opposed to the idea of evolution, but his studies of these objects convinced him that the earth was far older than people thought, and that the geological evidence showed that species had changed. It was in 1800 that he delivered a lecture in which he championed his theory of transformationism (transmutation), and in 1809 he published his greatest work, Philosophie Zoologique, in which this theory was elaborated.
Lamarck’s achievements are remarkable:
- He was one of the first to use the term ‘Biology’ to describe the subject. He argued that living organisms had properties that were unique but that these were the outcome (today we would say emergent properties) of the physics and chemistry of matter. He was a materialist, but one who realised that the characteristic of living organisms was their organisation and structure, not just their components.
- He thought that all organisms had developed from tiny sea creatures. He therefore had a version of the common ancestor idea.
- He saw that there was a direction in the evolutionary process towards greater complexity. This led him to the concept of ‘le pouvoir de la vie’ (the force of life).
There were precursors of Lamarck, just as Lamarck himself was a precursor of Darwin. But Lamarck should be recognised as one of the key scientists in the development of evolutionary biology. Darwin fully recognised this status: “this justly celebrated naturalist….who upholds the doctrine that all species, including man, are descended from other species.” (Preface to the 4th edition of The Origin of Species, 1866).
Several developments led to the blackening of his name.
- He championed, but did not invent, the idea that acquired characteristics could be inherited. That itself would have been fine. Darwin also accepted this idea. The problem was that the subsequent establishment of the Weismann barrier (isolation of the germ line) and then the Central Dogma (isolation of the genome) led to the idea being so completely ridiculed that Lamarck’s reputation was ruined.
- ‘Le pouvoir de la vie’ was often interpreted to mean belief in a special ‘vital force’. My reading of his work is that he certainly did not intend this interpretation. He was a materialist, not a vitalist.
- His great rival, Georges Cuvier, opposed the idea of evolution and wrote a devastating criticism of Lamarck, which was used at his funeral.
http://www.victorianweb.org/science/science_texts/cuvier/cuvier_on_lamarck.htm
The historian of Science, Pietro Corsi, has created a superb on-line resource on Lamarck:
I wrote an imagined letter from Lamarck:
http://musicoflife.website/pdfs/LetterfromLamarck.pdf
which expresses the reasons why I think he has been badly treated.
These books by Eva Jablonka and her co-authors/editors are also valuable resources, particularly on the various mechanisms by which ‘lamarckian’ inheritance occurs:
Jablonka, E. and M. Lamb (1995). Epigenetic inheritance and evolution. The Lamarckian dimension. Oxford, OUP.
Jablonka, E. and M. Lamb (2005). Evolution in Four Dimensions. Boston, MIT Press.
Gissis, S. B. and E. Jablonka, Eds. (2011). Transformations of Lamarckism. From Subtle Fluids to Molecular Biology. Cambridge, Mass, MIT Press.
Are genes followers rather than leaders in evolution?
This is the central question raised in an article in the New Scientist (12th October 2013). The article quotes Richard Dawkins: “which elements have the property that variations in them are replicated with the type of fidelity that potentially carries them through an indefinitely large number of evolutionary generations?” Dawkins answers his own question: “Genes certainly meet the criterion. If anything else does, let’s hear it.”
Nature is replying to that challenge loud and clear.
First, there are increasing numbers of experiments showing robust transgenerational inheritance independent of DNA. See Transgenerational inheritance.
Second, intricate cellular structure, static and dynamic, replicates itself faithfully and indefinitely through the process of self-templating. See Immortal Genes.
Experiments on cross-species cloning show the specific nature of this inheritance and how it determines the way in which the genome is interpreted.
Third, the famed ‘immortality’ of DNA is actually a property of cells. Cells have the machinery to correct frequent faults in DNA replication. It is also an elementary philosophical mistake to regard DNA as an active cause. On its own, DNA does nothing. See What does DNA do?
Finally, since DNA is not an active cause, it must be a follower in evolution. It is organisms that live or die and can therefore be subject to selection.
Those who are trapped inside the misleading 20th century discourse of genes being responsible for everything should read chapter 1 of The Music of Life, where it is shown that there could be no biological experiment to demonstrate attributes of selfishness or cooperation in DNA. The same point is made in depth in an article published in 2011. The central problem with The Selfish Gene is that it is unfalsifiable. See What is wrong with The Selfish Gene?
Randomness and Functional change
Defining what is meant by ‘random’ is itself a major field of enquiry in mathematics, computation and in science generally. The question whether there are truly random events in the universe is a vexed one, lying at the heart of theories of quantum mechanics. Probably, we will never know, perhaps cannot know in principle, the answer to that kind of question. But it warns us that defining randomness is not easy. I also say that in the lectures.
The best way to sidestep the deeper problems is to ask the question ‘random with respect to what?’ In evolutionary theory that makes the problem much simpler. Both Neo-darwinists and their opponents can then agree that what is really meant is ‘random with respect to physiological (phenotypic) function’. That is so because one of the central tenets of Neo-darwinism is the exclusion of any form of Lamarckism, the idea that function, or functional improvement, can influence inheritance, i.e. the inheritance of acquired characteristics. By contrast, a Lamarckian must maintain that at least some changes are not completely random with respect to function.
In the article, and inevitably in much shorter form in the lectures, I approach this question in three stages. The first stage is to establish that genomic change is not random with respect to location in the genome. The reason for asking that question first is that without establishing that there are preferred locations of change, the argument for any kind of functionally-relevant change cannot even get off the ground. The only way in which such a change can occur is through influencing the physical and chemical properties of the genetic material. Preferred locations of change are therefore a pre-condition for functional change to be possible. If all locations in the genome were equipotent for changes there would be no possibility for functionally-relevant change.
Of course, demonstrating the existence of hotspots and other ways in which change is not randomly distributed with respect to location does not, in and of itself, demonstrate any form of functionally-relevant change. The existence of hotspots could be simply a consequence of the physico-chemical properties of the genome and its associated proteins even if no functionally-relevant changes occur. Further experimentation is required to demonstrate that. I also make that clear in the article and lectures.
The second stage in the argument is to note that well-documented examples of functionally-relevant genomic change already exist. The best-investigated case is the evolution of lymphocytes. In response to antigen activation the relevant part of the genome undergoes very rapid proliferation. Targeted speeding-up of change is therefore one mechanism by which functional change can occur. That is true even if the individual changes at that location are random. The functionality lies in the targeting of the location. A similar targeting occurs in P element homing. And targeted reorganisation of the genome by speeding up change in selected locations occurs in bacteria subject to starvation. All these mechanisms are referred to in the item Relevance to physiology.
The third stage in the argument is experimental demonstration that the inheritance of acquired characteristics occurs. There are now many examples of that. See Transgenerational Inheritance. Those experimental results require functional inherited change, either genetically or epigenetically, even if we do not yet know the molecular mechanisms. All these stages of the argument are necessary. Some of the critics have mistaken the first step for the whole argument!
Links to video lectures and the Sourcebook
(100 pages – all the evidence and references – free of charge)
Population heritability estimates
Contributed by Michael Joyner
In the 1800s biometrics, the measurement and classification of human phenotype, emerged as a field of study. Early biometric studies showed for example that parental height could account for about 40% of the variance seen in children. As ideas about the biological basis of both between and within species phenotypic variability emerged post-Darwin, there has been a long lasting effort to determine the biological basis of heritability. This accelerated in the early 1900s with the rediscovery of Mendel’s work on clearly heritable phenotypic variation in plants. It also led to the coining of the terms gene, genotype and phenotype, and also the discovery of chromosomes as the cellular home of “genes”. The idea that phenotypic variability had a clear cut and mostly genetic explanation continued as the DNA centric view of “what is a gene” emerged in the 1950s. The shift in the definition of a gene also appears to have contributed to the idea that variability in DNA sequences would be the major driver of phenotypic variability. In this context, a frequently overlook concept is that estimates of heritability are also dependent on relatively stable environmental and cultural conditions. For example heritability estimates for body mass index (BMI, a measurement linked to body composition and used in the study of obesity) are dependent on both the socio economic status of the population being studied and also any recent changes in the socio economic status of the population being studied. Likewise in Japan heritability estimates for height since World War II are confounded by large generational increases in height due to improved nutrition in the absence of changes in DNA sequence. For many human phenotypes including the prevalence of many non-communicable diseases, migration studies show that environment, behavior and cultural factors play a major role in human phenotypic variation in the absence of DNA sequence differences. Observations such as these highlight the limitations of the DNA centric view of human phenotypic variation. They emphasize the idea that complex multidirectional interactions between the environment, behavior, physiological regulation and adaptation, and the genome explain human phenotypic variation.
For more information see:
Aulchenko YS et al (2009). Predicting human height by Victorian and genomic methods. Aur J Hum Genet 17: 1070-1075. http://www.nature.com/ejhg/journal/v17/n8/full/ejhg20095a.html
Fisher RA (1919). The causes of human variability. Eugene Rev 10: 213-220. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2942138/
Johannsen W (1911). The genotype conception of heredity. Am Naturalist XLV: 129-159. http://www.jstor.org/stable/2455747?__redirected
Joyner MJ, Prendergast FG (2014). Chasing Mendel: five questions for personalized medicine. J. Physiol in Press.
Marmot MG, Syme SL (1976). Acculturation and coronary heart disease in Japanese-Americans. Am J Epidemiol 104: 225-247. http://www.ncbi.nlm.nih.gov/pubmed/961690



